PAVE User's Guide - Version 3.0
- Introduction
- Download and Installation
- Types of plots produced by PAVE
- Getting your data into PAVE
- Using formulas
- Spatial and temporal data subsetting
- Navigating through PAVE's menu items
- Configuring plots
- Printing and exporting images, animations, and data
- Driving PAVE using scripts
- Optional Environment Variables
- Requirements for use
- Quick PAVE Jumpstart
- Known bugs and workarounds
- Run time errors
- History of new features
- PAVE FAQ
- Future PAVE Development
- Return To cjcoats.github.io
NOTE As of the release for pave-3.0,
much of this document is obsolete (as much as 20 years out of date),
and the original wasa full of HTML-errors; however it is a tedious
and detailed task to bring it fully up to date.
For usage-changes (and particularly new capabilities in the
wrapper-script), see the document ${PAVE_DIR}/AAREEADME
-- Carlie J. Coats, Jr., Ph.D.
pave-3.0 maintainer.
This document describes how to use the Package for Analysis and
Visualization of Environmental data (PAVE). PAVE is a flexible
application to visualize multivariate gridded environmental datasets.
Features include
- baseline graphics with the option to export data to high-end
commercial packages,
- access and manipulation of datasets located on remote
machines,
- support for multiple simultaneous visualizations,
- an architecture that allows PAVE to be controlled by external
processes,
- low computational overhead, and
- no software distribution cost.
PAVE is licensed under the terms of the GNU Public License
Version 2 copyright © 1989, 1991 Free Software Foundation, Inc.
See its enclosed GPL.txt or
https://www.gnu.org/licenses/old-licenses/gpl-2.0.en.html.
PAVE Version 3.0 is being released on GitHub in
March 2018. There are multiple changes from Version 2.3:
- Linux Medium-memory-model support: File-size, etc., are
essentially limited by available RAM and inherent netCDF
limitations (no more than 2 GB per time step). See
https://cjcoats.github.io/ioapi/AVAIL.html#medium
- The primary build is for GNU gcc/g++/gfortran compilers, using
I/O API build type
BIN=Linux2_x86_64gfort_medium; the build
is "as static as possible" and hopefully should run on most
64-bit Linux systems "as is." The rest of these systems
should need minor Makefile customization, followed by
make relink.
- Major restructuring as a stand-alone application. References
to, and the convoluted directory structures from, MCNC's
EDSS have been removed. Also, additional maps were added,
and placed in (build-independdent!) directory
PAVE_MAPDIR = pave-3.0/maps/.
- The driver-script
PAVE_DIR/bin/pave was
extensively re-written, offers additional capabilities,
and is much more easily customized than its predecessor.
- PAVE documentation is included as part of the package, in
directory
PAVE_DIR/Docs/ Note that much of the
original documentation fails the weblint
HTML-validation test—badly!—and ios taking some
not-inconsiderable time and effort to fix.
Frankly, the documents are badly obsolete and need a lot of
work ;-( This is a work-in-progress...
- Provided compressed source-code tar-balls for (most of) the
external libraries needed by PAVE, together with scripts for
uncompressing, configuring, building, and installing them,
compatibly with the primary
Linux2_x86_64gfort_medium binary build type, for
re-building as necessary (the installed libraries, etc., are
already in the distributed package's
${PAVE_DIR}/bin and ${PAVE_DIR}/lib)
See ${PAVE_DIR}/INSTALL for instructions on
re-building PAVE.
- Many loop-restructuring and other optimizations: in previous
versions, loop-nests over grids were basically structured as
badly as possible; grid-statistics, formulas, etc., should
be at least an order of magnitude faster now (the larger the
dataset, the larger the speedup).
- Support for new map-projections (not fully tested yet...):
Albers Equal Area
Polar Stereograph
Equatorial Mercator
(General) Transverse Mercator
Lambert Azimuthal Equal Area
- Map-drawing capabilities were added, to draw maps for
"extended" regions that cross the International Date
Line, or that follow the WMO's so-called "Longitude"
definition that contradicts the International Standards
Organization's Standard 6709.
- Fixed a number of "grid hacks" in
pave/utils.c
that assume whenever I/O API
XCELL3D < 100M, the file is a (bad,
1993-vintage?) mis-formed file where the user has mistakenly
described the grid in Km units instead of M. These hacks in
the original destroy the usability of PAVE for high resolution
hydrological-modeling grids.
- Re-formatted the code for consistency and readability, using
astyle --style=whitesmith --indent=spaces=4 \
--indent-col1-comments --pad-paren --convert-tabs
PAVE version 2.3 was released on October 18, 2004; the previous
public release of PAVE was version 2.2 in June, 2004.
See New Features
for more information on the improvements made in these and
earlier versions.
We hope you enjoy your PAVE visualizing experience!
On-line PAVE Resources
- Download PAVE
-
git clone https://github.com/cjcoats/pave-3.0
- PAVE Frequently Asked Questions
- Getting started with PAVE
Return To Table of Contents

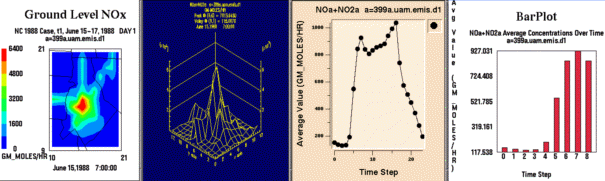
From left to right, these are examples of a smoothed tile plot, a
3D mesh plot, a time series line plot, and a time series bar plot.

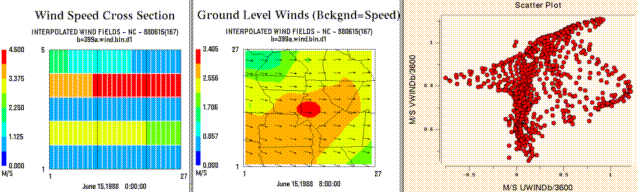
From left to right, these are examples of a tile plot of a vertical
cross section, a wind vector plot, and a scatter plot.
Return To Table of Contents
PAVE currently supports four input file formats:
(which contain a list files of one of the other three types). Files
of these types are loaded into PAVE from the Add/Delete/Select
Dataset_popup window. This window comes up automatically when
you start PAVE, and also appears when you choose Edit/Select From
Dataset List from PAVE's Datasets menu. See
Quick PAVE Jumpstart for more details on
loading datasets.
I/O API Data
The EPA/MCNC/BAMS/IE
Models-3 Input/Output Applications Programming Interface (see
https:cjcoats.github.io/ioapi/AA.html for further info) provides
an easy-to-learn, easy-to-use programming interface to files for the
air quality model developer and model-related-tool developer, in
both Fortran and C/C++. I/O API files are portable across
computing platforms. This means that the same file can be read on a
PC, a Sun workstation, a DEC Alpha workstation, and on a Cray
supercomputer. If your data is not already in I/O API, UAM-IV,
or UAM-V format,formats we recommend that you use a translation
program such as those found in the I/O API's
m3tools package to convert your data into I/O API
format. Note that these latter are available in source code form,
so that you may adapt them easily to your needs. See in particular
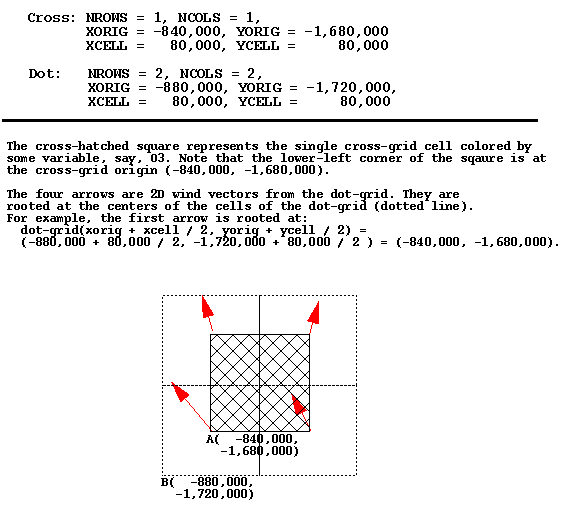
One sometimes confusing concept is the precise meaning of the
XORIG, YORIG, XCELL, and
YCELL parameters in an I/O API data file. See
https:cjcoats.github.io/ioapi//GRIDS.html for the I/O API
documentation on this subject. Below is an image of an example of
using these parameters with two different data files that need to be
co-registered within a PAVE plot. Note that units are in meters,
except in the case of files with Lat-Lon coordinates, where they are
in (signed) degrees.
UAM-IV Format
Most geographically oriented
UAM-IV input and output files are directly readable by PAVE. These include
DIFFBREAK, REGIONTOP, TEMPERATURE, WIND, AIRQUALITY, BOUNDARY, TOPCONC,
EMISSIONS, PTSOURCE, AVERAGE, and INSTANT.
UAM-V Format
Some of the UAM-V input and output files are compatible with the UAM-IV
file formats. These files are readily visualized by PAVE because it can
obtain all the information it needs from inside the files (i.e. they are
"self-describing"). The files in this category are: EMISSIONS, PTSOURCE,
BOUNDARY, AIRQUALITY, and the coarse grid AVERAGE and INSTANT files.
However, many UAM-V files have new formats that are not self-describing.
PAVE needs additional information in order to read these files, such as the
kind of data in the file, the number of rows, columns, and layers in the
data, the geographic region covered, etc. The file types that fall into
this category are wind, temperature, cloud, water vapor, rain, vertical
diffusion, height, fine grid average, and fine grid instant.
In order to display a file of one of these latter types with PAVE, a
UAM-V "metafile" must be used. A metafile is an ASCII file that contains
the additional information PAVE needs to read and correctly interpret the
data. To visualize data in a file that needs a metafile, select the
metafile from the PAVE file browser, instead of the file that contains
the data. A description of the contents of UAM-V metafiles follows.
The very first line of the metafile MUST be the following:
#! UAMV DESCRIPTION FILE
If PAVE does not find the above string, the file is considered to be
of unknown type and an error is returned.
Several keywords must be present in the metafile. Each keyword
should be on a separate line and followed by a value. Blank lines
are permitted. The following keywords are required (and are listed
in the recommended order):
UAMV_FILE
UAMV_TYPE
NCOLS
NROWS
LEVELS
XORG
YORG
DX or DLON
DY or DLAT
UTM_ZONE (for UTM-based domains only)
FINE_GRID (for UAM-V output files only - not for input meteorology files)
TITLE
Keyword Descriptions
UAMV_FILE - the name of the file that contains the actual data
to be displayed by PAVE. It can be either a full or a relative pathname to
the current working directory.
UAMV_TYPE - the type of data in the UAMV_FILE. Valid choices are:
Wind, Temp, Cloud, H2O, Rain, Vdif, Height, FineGridAverage,
FineGridInstant
NCOLS - number of horizontal columns in the grid
NROWS - number of horizontal rows in the grid
LEVELS - number of vertical layers in the grid
XORG - the x-coordinate of the lower left corner of the grid, in km for
UTM-based grids and degrees for latitude-longitude grids
YORG - the y-coordinate of the lower left corner of the grid, in km
for UTM-based grids and degrees for latitude-longitude grids
DX - size of the horizontal cell in the x-direction for
UTM-based grids (km)
DLON - size of the horizontal cell in x-direction for lat-lon grids
(degrees)
DY - size of the horizontal cell in y-direction for UTM-based grids
(km)
DLAT - size of the horizontal cell in y-direction for lat-lon grids
(degrees)
FINE_GRID - logical variable. Possible values: 0 and 1. If the value
is 1, the file represents data on a fine grid, otherwise it is on a coarse
grid. For meteorological input files, this value should be set to one only
if there is a layer of cells outside the domain specified by { XORG, YORG,
NROWS, NCOLS, DX, and DY]}.
TITLE - A title for tile plots. Spaces are allowed.
Example of a UAM-V temperature metafile on the OTAG coarse grid:
#! UAMV DESCRIPTION FILE
UAMV_FILE /home/user/tmpr.cc.20jul93.ld.rams1
UAMV_TYPE Temp
NCOLS 64
NROWS 63
LEVELS 5
XORG -99.0
YORG 26.0
DLON 0.5
DLAT 0.3333333
FINE_GRID 0
TITLE Coarse Grid Temperature: July 20, 1993
Example of a UAM-V fine grid average metafile on the OTAG fine grid:
#! UAMV DESCRIPTION FILE
UAMV_FILE /home/trayanov/testpave/avrg.ff.20jul93-93.mc.93basA1
UAMV_TYPE FineGridAverage
NCOLS 137
NROWS 110
LEVELS 7
XORG -92.0
YORG 32.0
DLON 0.16666667
DLAT 0.11111111
FINE_GRID 1
TITLE Base Case Fine Grid Average
Example of a UAM-V temperature file on a UTM-based grid:
#! UAMV DESCRIPTION FILE
UAMV_FILE /metdata/uamv.t.jun26-28.16km.8.v4
UAMV_TYPE Temp
NCOLS 35
NROWS 50
LEVELS 8
XORG 200.0
YORG 4400.0
DX 16.0
DY 16.0
UTM_ZONE 16
FINE_GRID 0
TITLE Coarse Grid Temperature File
Creating metafiles with scripts
It is relatively straightforward to use a script to create
metafiles. Examples of scripts that do this follow. However, the
specifics (such as parsing file names to determine file type) depend
on the particular application. For PAVE-3.0,There are two examples
make_in_metas and make_out_metas that were
originally created for the OTAG project; these can be found in
${PAVE_DIR}/bin/
Viewing multiple sequential files as one dataset
using chain files
Data from multiple files can be concatenated and displayed as if they
were from a single file. This is useful if you wish to animate over
sequential data that is stored in multiple files. In order to do this
you need to supply a "chain file" - an ASCII file that
contains a list of the data files to be concatenated.
The first line of a chain file must be
#! LIST_OF_CHAINED_FILES
If this is not found, PAVE will not recognize that the file is a chain file.
A list of full path names to the individual files to be concatenated
should follow this line with one filename per line. Note that no
blank lines or comment lines are permitted and the files MUST be in the order
in which you wish the data to be displayed. The individual files can be one
of the following types: netCDF, UAM-IV or UAM-V (regular or meta files).
A script is provided with PAVE to simplify the creation of chain files.
The script will currently work only in cases where the files to be
chained reside in a single directory. To create a chain file, cd to the
directory that contains the files you wish to chain, and type
chain_files name_of_chain_file list_of_files_to_chain
For example:
chain_files avrg.cc.20-30jul93-93.mc.basB avrg.cc.*
Limitations:
The current version of PAVE does not check the files for
consistency (i.e. whether they are of the same type, whether the grid is the
same in all the files, or whether the files are listed in the proper
sequential order).
Here is an example of a chain file:
#! LIST_OF_CHAINED_FILES
/home/trayanov/testpave/rain.cc.20jul93.ai.meta
/home/trayanov/testpave/rain.cc.21jul93.ai.meta
/home/trayanov/testpave/rain.cc.22jul93.ai.meta
Return To Table of Contents
One of PAVE's most powerful features is its formula capability,
which enables you to calculate and visualize derived variables from
your datasets "on the fly". For example, you can calculate
the ratio of a variable from one file to a variable from another
file, and then visualize the ratio. It is easy to load formulas
into PAVE using the Add/Delete/Select Formula_popup window,
which appears automatically when you start PAVE. The window can
also be brought up manually by choosing Edit/Select From Formula
List from PAVE's Formulas menu. See
Quick PAVE Jumpstart for more details on
loading formulas.
All PAVE visualizations are generated using one or more formulas. A
formula may be very simple. For example, the formula
O3a refers to the variable O3 in data set
a - which is the first dataset that was loaded into PAVE.
(Note that data sets are given sequential letters as they are loaded
into PAVE, and are referred to by those letters in PAVE formulas.)
An example of a formula to calculate the percent difference in
O3 between datasets a and b is:
(O3a-O3b)*100/(O3b+0.00001).
Formulas must be in infix notation, and can contain the following operators,
listed in their order of precedence:
Highest 1) abs, sqr, sqrt, exp, log, ln, sin, cos, tan,
Precedence sind, cosd, tand, minx, miny, minz, maxx, maxy, maxz,
mint, maxt, mean, sum, min, max,
2) **
3) /, *
4) +, -
5) <, <=, > >=
6) ==, !=
7) &&
Lowest 8) ||
Precedence
Explanations of these operations are given below. If you wish to
override the default operator precedence, or are uncertain as to
which operator will take precedence, you can feel free to use
parentheses in your formulas. This will force expressions within
the parentheses to be evaluated first.
PAVE also has an occasionally used feature that allows you to
specify a time step index after a CODEiable name. For example,
O3a:1 is the first hour of ozone. So, if you
wanted to plot each cell averaged in time over the first six hours
of your data, you could enter and plot the following formula:
(O3a:1+O3a:2+O3a:3+O3a:4+O3a:5+O3a:6)/6
This is cumbersome and it also uses a lot of memory, but it may be
useful for you.
There is another useful feature of the parser that not many people
know about, that enables you to compute and visualize the the
rate of change of a variable. For example, the formula
d[O3a]/dt calculates the change in ozone concentration
over time. A limitation of this feature is that the variable
between the brackets must be an atomic variable, that is to
say, it can not be a formula other than a basic variable from one of
your datasets.
Formulas may also contain integer or floating point constants, or
the following operands which are replaced by PAVE's formula parser
to be the constant values noted:
- E
- 2.7182818284590452354
- PI
- 3.14159265358979323846
- NROWS
- number of rows in the formula's currently selected domain
- NCOLS
- number of rows in the formula's currently selected domain
- NLEVELS
- number of levels in the formula's currently selected domain
The following operators are binary (they have an operand on both
sides of the operator), and usually return an array of data by
performing that operation on each cell of the operands' arrays. The
only time these operators return a single number is when both
operands (ops) are themselves a single number.
+
- Returns the sum of the operands
-
- Returns the difference of the operands
*
- Returns the product of the operands
/
- Returns ratio of the operands
**
- Returns the left operand raised to the power of the
right operand, calculated using the C math library's
pow() function
The following operators are boolean binary operators. Boolean operators
return either 0 or 1 for each cell of the resulting array of data, or
in the case of two operands (ops) that are single numbers, just the single
number 0 or 1. You may find these
operators useful to "screen out" ranges of your data that are
of particular interest. For example, if you are only
concerned about the variable O3a when its value exceeds 0.080, you
might look at the formula (O3a>0.080)*O3a. If O3a
is less than or equal to 0.080, the result of the formula will
be set to 0 in that cell. Otherwise, the value of O3a will be
used in that cell.
<
- Returns 1 if the left op is less than the right op,
<=
- Returns 1 if the left op is less than or equal to
the right op, else 0
>
- Returns 1 if the left op is greater than the right
op, else 0
>=
- Returns 1 if the left op is greater than or equal
to the right op, else 0
!=
- Returns 1 if the left op is not equal to the
right op, else 0
==
- Returns 1 if the left op is equal to the right op, else 0
&&
- Returns 1 if both ops are non-zero, else 0
||
- Returns 1 if either op is non-zero, else 0
The following operators are unary (they have a single operand on the
right side of the operator), and usually return a time-stepped matrix of
data by performing that operation on each cell of the operand's array.
The only time these operators return a single number is when the operand
is itself a single number. The C math library routines called are listed
with most of these operators. For further information on these routines,
please check your man pages.
- abs
- fabs(op)
- sqrt
- sqrt(op)
- sqr
- Returns the square of the op
- log
- log10(op)
- exp
- exp(op)
- ln
- log(op)
- sin
- sin(op)
- cos
- cos(op)
- tan
- tan(op)
- sind
- sin(op*(PI/180.0))
- cosd
- cos(op*(PI/180.0))
- tand
- tan(op*(PI/180.0))
The following unary operators return a single number in all cases.
Their single operand must follow on the right hand side of the
operator. The functionality is listed beside each operator name.
- mean
- average cell value for all cells in currently selected domain
- sum
- sum of all cell values in currently selected domain
- mint
- time step index with minimum value in currently selected domain
- maxt
- time step index with maximum value in currently selected domain
- minx
-
X index with minimum value in currently selected domain
- maxx
-
X index with maximum value in currently selected domain
- miny
-
Y index with minimum value in currently selected domain
- maxy
-
Y index with maximum value in currently selected domain
- minz
-
Z index with minimum value in currently selected domain
- maxz
-
Z index with maximum value in currently selected domain
- nrows
- number of rows in the currently selected domain
- ncols
- number of rows in the currently selected domain
- nlevels
- number of levels in the currently selected domain
where "currently selected domain" includes the currently
selected rows, columns, layers, and time steps. So the
currently selected domain is bounded by
(minx,miny,minz,mint)<->(maxx,maxy,maxz,maxt)
The unary min and max operators behave a little differently:
- min
- For each cell (i,j,k) in the currently selected domain,
this calculates the minimum value for that cell
over the currently selected time steps. In other words,
the minimum value in cells (i,j,k,tmin..tmax).
- max
- For each cell (i,j,k) in the currently selected domain,
this calculates the maximum value for that cell
over the currently selected time steps. In other words,
the maximum value in cells (i,j,k,tmin..tmax).
NOTE: currently the unary + and - operators [as in -1 or -(x+y)]
are not supported, but hopefully these will be added later.
Return To Table of Contents
PAVE allows you to easily subset your data by geographic region,
layer range, and time range. This section explains the concepts of
how PAVE manages this information in its memory space. An
understanding of this section should help you with your PAVE usage.
To adjust layer ranges, geographic regions of interest (domains), or
time step ranges for datasets and formulas, you can use menu items
in the Datasets and Formulas menus on PAVE's main
interface window.
]
Each dataset has some number of layers greater than or equal to one,
some number of time steps greater than or equal to one, and some
geographic region onto which its grid maps. A geographic region,
often referred to as a domain, is defined by the map projection
type (Lat/Lon, UTM, Lambert Conformal, etc.), the number of rows and
columns in the dataset, and the geographic boundaries of the area on
which the grid falls.
For each unique geographic region (or domain) that PAVE learns about
by examining the dataset(s) chosen by the user, PAVE keeps a single
domain object. Domain objects are where the currently
selected geographic region for formulas and datasets are stored in
memory. When a domain object is created by PAVE, it defaults to
having all of its cells selected, until the user chooses a subregion
within that domain. Because PAVE only stores a single domain object
for each unique domain that it learns of, there may be numerous
datasets and formulas that make use of the same domain object.
Therefore, when a user chooses to modify a formula's or a
dataset's selected region of interest, the region of interest for
all formulas and datasets that refer to the same region and
have the same grid dimensions will be affected. Any subsequent plots
of any formulas who rely on that domain object will reflect its
newly selected geographic region of interest.
Similarly, PAVE uses layers objects to store information
about currently selected layer ranges. Each dataset i has some Ni
number of layers associated with it, and each formula j has some Nj
number of layers associated with it. For each unique number of
layers that PAVE learns about, PAVE keeps a single layers
object in memory. Therefore, when a user chooses to modify a
formula's or a dataset's selected layer range, there may be
more than one dataset and more than one formula on which the
modified layer range has an effect. When PAVE learns about a new
unique number of layers, the layers object that it creates by
default has only the first layer selected, until the user modifies
the layer range for this layers object.
Time ranges are handled differently than layer ranges and geographic
regions of interest. Each dataset has exactly one unique time
range object associated with it. That time range object is
associated only with that one dataset. Each formula also has
exactly one unique time range object associated with it. That time
range object is associated only with that one formula. Because a
formula may rely on more than one dataset to derive its data, a
formula's time step range is always bounded by the time ranges of
the datasets it depends on. For example, a formula could have
three datasets from which it derives data. Those datasets could
have 72, 48, and 24 time steps of data in their currently selected
time range objects. In this case, the maximum number of time steps
in that formula's time range object could ever have would be 24.
Should the user reduce the number of steps in the currently
selected time range of any of the three datasets to less than 24,
then the maximum number of time steps in that formula's time range
object would immediately be reduced accordingly.
Return To Table of
Contents
PAVE's main graphical user interface is broken into six menus:
This section briefly describes each of the items on these menus.
The file menu has six menu items.
Choose Configuration File For New Tile Plots brings up a file
browser that allows you to select a file that contains configuation
settings for tile plots. Settings can include information such as colors
to use, number of colors to use, min and max values for the legend, etc.
These settings will be used for all subsequent tile plots, until another
configuration file is chosen. The ASCII configuration files can be
edited manually, or saved directly from a tile plot. For further
information, please see the section on
Configuring plots.
Load Dataset List From File brings up a file browser that
allows you to select an ASCII file that contains a list of datasets that
you would like to visualize with PAVE. Loading in a dataset list from a
file will first cause PAVE to remove any currently loaded datasets from
PAVE's memory. In this way, dataset names loaded in will then be
referenced by letters a, b, c, etc.
Load Formula List From File brings up a file browser that
allows you to select an ASCII file that contains a list of formulas that
you would like to visualize with PAVE. Note that any formulas loaded in
will be verified against the currently loaded datasets. As long as the
variables in the formulas match variables in the respective datasets,
they will be loaded into PAVE. Otherwise they will be ignored.
Save Dataset List To File allows you to save a list of the
currently loaded datasets to an ASCII file. If you want to look at the
same set of datasets in many different PAVE sessions, you may find
this feature useful. Dataset lists can subsequently be retrieved
using the Load Dataset List From File menu item described above.
Save Formula List To File allows you to save a list of the
currently loaded formulas to an ASCII file. If you want to look at the
same set of formulas in many different PAVE sessions, you may find it
useful to use this feature. Formula lists can subsequently be retrieved
using the Load Formula List From File menu item described above.
Exit PAVE causes your PAVE session to end.
The datasets menu allows you to modify the information PAVE stores
regarding available datasets.
Edit/Select From Dataset List brings up an "Add/Delete/Select
Dataset" window that allows to you load and delete datasets from
PAVE's memory. Click on the "Add" button, and an EDSS File Browser
should appear (this may take a couple of seconds). You can then browse
for datasets on your local machine and on remote machines if your system
is properly configured. For more information on using the EDSS File
Browser, see Quick Pave Jumpstart.
If you want to browse for files on remote machines, first verify that
you've met the requirements discussed in the
Requirements for use
section of this document. Then click on the
large button in the center of the EDSS File Browser that contains
information about "Host:", "User:", and "Owner Module:". This brings up a
"host selector" window in which you can enter the remote host name (e.g.
sequoia.nesc.epa.gov). Clicking on "Select" will cause the file browser
to go to your home directory on the remote machine if your system is
configured properly. You should then be able to browse for files on that
machine.
Datasets selected with the EDSS File Browser are loaded into PAVE
sequentially, and are referenced using the letters a, b,
c, etc.
When a new dataset it loaded into PAVE, it becomes the currently
selected dataset. You can always determine which is the currently
selected dataset by looking at the "Dataset:" line on the main
PAVE window. You can change the currently selected dataset
by simply single-clicking on the desired dataset name in the
"Add/Delete/Select Dataset" window. PAVE's "Species List"
window will then contain the names of the variables contained in the
dataset, each followed by the letter corresponding to the dataset.
Clicking on a dataset name and then clicking on the "Delete" button will
remove that dataset from PAVE's memory. Clicking on the "Close" button
will close the window.
Select Time Range of Current Dataset brings up a window
with two sliders that can be used to crop the currently selected
dataset's time range to a smaller time range, which by default is set to
the maximum range in the dataset. Cropping, or subselecting, a dataset's
time range will affect any plots made using that dataset's variables.
The maximum time range that can be used for any of the variables in a
plot is specified by these sliders. Please see the section on
Spatial and temporal data subsetting
for further information on how PAVE stores time ranges.
Select Layer Ranges Matching Current Dataset brings up a window
with two sliders that can be used to change the currently selected
dataset's layer range to a different layer range. By default, a dataset's
layer range is set to be layer 1 (ground level) only. Changing a
dataset's layer range will affect any plots made using that dataset's
variables, and may also affect plots made using other datasets. The
maximum layer range that can be used for any of the variables in a plot
is specified by these sliders. Note that there may be multiple datasets
and formulas that share the same layer range information. Please see the
section on Spatial and temporal data
subsetting for further information on how PAVE stores layer ranges.
Select Regions Of Interest Matching Current Dataset brings up a
window that can be used to change the currently selected dataset's
geographic region of interest. By default, a dataset's geographic region
of interest is the entire region referenced by the dataset. This window
can be used to select part of this region. Changing a dataset's
geographic region of interest will affect any plots made using that
dataset's variables, and possibly those of other data sets because there
may be multiple datasets and formulas that refer to the same region of
interest.
The geographic region of interest window has its own menu. The File menu
can be used to save a geographic region of interest to a file, retrieve a
region of interest from a file, or to close the window. The Edit menu
can be used to easily select all of the grid cells in the window, or to
select none of them. Cells can explicitly be toggled on or off by
dragging the mouse over the region to be toggled. You may find that
selecting a particular region is easier if you resize the window to make
it larger. Please see the section on Spatial
and temporal data subsetting for further information on how PAVE
stores geographic regions of interest.
View Variables In Current Dataset will cause a window to appear
that lists the variables in the currently selected dataset, followed by
the letter that corresponds to the dataset. You can click on any of the
variables to cause that variable to be added to the formula list and
become the currently selected formula. The formula can then be plotted
using selections from the Graphics menu without any further clicks of the
mouse.
The formulas menu allows you to modify the information PAVE stores
regarding available formulas.
Edit/Select From Formula List brings up an "Add/Delete/Select
Formula" window that allows to you add and delete formulas from PAVE's
memory. You can enter a formula using the "Enter New Formula:" typein
widget according to the rules described in the
Using formulas section, then click on the Add button to load it
into PAVE's memory.
When a new formula is loaded into PAVE, it becomes the currently selected
formula. You can always determine which is the currently selected
formula by looking at the "Formula:" line on the main PAVE control
window. You can change the currently selected formula by simply clicking
on the desired formula name in the "Add/Delete/Select Formula" window.
Clicking on a formula and then clicking on the "Delete"
button will remove that formula from PAVE's memory.
Clicking on the "Close" button will close the window.
Select Time Range of Current Formula brings up a window
with two sliders that can be used to crop the currently selected
formula's time range to a smaller time range. Cropping, or
subselecting, a formula's time range will affect any plots made using
that formula. The maximum time range that can be used for the formula
in a plot is specified by these sliders. Please see the section on Spatial and temporal data subsetting for further
information on how PAVE stores time ranges.
Select Layer Ranges Matching Current Formula brings up a window
with two sliders that can be used to change the currently selected
formula's layer range to a different layer range. By default, a formula's
layer range is set to be layer 1 (ground level) only. Changing a
formula's layer range will affect any plots made using that formula, and
possibly that of other plots too since there may be multiple datasets and
formulas that share this same layer range information. Please see the
section on Spatial and temporal data
subsetting for further information on how PAVE stores layer ranges.
Select Regions Of Interest Matching Current Formula brings up a
window that can be used to change the currently selected formula's
geographic region of interest. By default, a formula's geographic region
of interest is set to be the entire region referenced by the formula .
This window can be used to select part of this region. Changing a
formula's geographic region of interest will affect any plots made using
that formula's variables, and possibly other plots since there may be
multiple datasets and formulas that share this same geographic region of
interest information.
The geographic region of interest window has its own menu. The File menu
can be used to save a geographic region of interest to a file, retrieve a
region of interest from a file, or to close the window. The Edit menu
can be used to easily select all of the grid cells on the window, or to
select none of them. Cells can explicitly be toggled on or off by
dragging the mouse over the region to be toggled. You may find that
selecting a particular region is easier if you resize the window to make
it larger.
Please see the section on Spatial and temporal
data subsetting for further information on how PAVE stores geographic
regions of interest.
The graphics menu is used to create plots.
Create Tile Plot makes a tile plot of data for the currently
selected formula using that formula's selected geographic region of
interest, layer range, and time step range. A tile plot has its own
menus that can be used to operate on that tile plot and/or its data.
- The tile plot's File menu can be used to print and save images
and animations; these are described more thoroughly in the
Printing and exporting images,
animations, and data section. Also this menu has a
"Save Configuration Settings" item that saves the tile plots legend
settings to a file, as discussed in the
Saving configuration settings for later use subsection of the
Configuring plots section.
- The tile plot's Interact menu can be used to change the effect of
a dragging the mouse pointer to select a portion of the tile plot's
cells. Interact..Probe..Tiles causes dragging to bring up a window with
the raw data values for selected region. Interact..Probe..Obs causes dragging to
bring up a window with the raw data values for observational data in the selected region.
(this option is greyed out if no observational data has been loaded using the -obs option)
Interact..Zoom causes dragging to zoom in on the selected region.
Interact..Time Series..Tiles causes dragging to create a time series
line plot of the average values over the selected region. Interact..Time
Series..Obs causes dragging to create a time series line plot of the
average values of the observational data over the selected region.
(this option is greyed out if no observational data has been loaded using the -obs option)
- The tile plot's Control menu can be used to bring up several
windows. The Control..Animate.. item displays a window that can be used
to animate through the tile plot's time steps. The Control..Zoom.. item
displays a window that is used to switch between all the different Zoom
regions that have been selected for this plot. The Control..Configure..
item brings up the tile plot's configuration window, discussed in detail
in the Configuring plots section of this
document.
- The tile plot's Map menu is used to toggle between five different
map overlays. The following maps are built into PAVE: Counties,
Medium-res States, High-res States, Rivers, Roads/transport routes,
and a world map. If the world map is used for a region that is
not within the North American continent, you will probably need to
adjust PAVE's "preclip" region using the preclip command line argument or
its corresponding environment variables. Otherwise your map will be
limited to lines within North America.
Note that for a given map projection, PAVE may be quite slow the first
time it displays a given map within a session. This is because PAVE
calculates the map on-the-fly with respect to the map-projection of the
dataset. Subsequent tile plots that use the same map and projection
will be faster, since PAVE caches map projections in memory for future
use.
- The tile plot's Plot menu can be used to create a time series line
plot at the cell with the maximum or minimum value in that tile plot,
over the entire time range of that tile plot.
- The tile plot's Overlay menu can be used to overlay observational
data.The Overlay..Observations..item displays an overlay popup window that is
used to select the observation data that will be overlayed on the existing
tile plot.
The Overlay..VectorObs.. item displays a vectorPlotObs popup window that
is used to select the formulas for the two components of the vector to
be overlayed on the tile plot.
The Overlay..Contour.... item displays an overlay_popup window that is
used to select the formula that will be overlayed as a contour
Create Vector Plot makes a vector plot from the formula selected
as component 1 and the formula selected as component 2.
The formulas selected as the components of the vector must match
in geographic region of interest, layer range, and time step range.
- The VectorPlot popup window is used by the user to specify the formula
for component 1 by clicking on Component 1, then clicking on a formula in
the Available Formulas window, and then clicking on Apply. The user then
clicks on Component 2, then clicks on a formula in the Available Formulas window, and
then clicking on apply again. After the components have been selected using
this method, the user may click on Components 1 and 2 to view the formula selected for
each component. Once the user has verified that the correct components are selected, then
click OK, and the Vector Plot will be displayed.
Create Vector Tile Plot overlays a vector plot from the formula selected
as component 1 and the formula selected as component 2 with a tile plot of a scalar variable.
The formulas selected as the components of the vector, and the scalar must match
in geographic region of interest, layer range, and time step range.
- The VectorTilePlot popup window is used by the user to specify the formula
for component 1 by clicking on Component 1, then clicking on a formula in
the Available Formulas window, and then clicking on Apply. The user then
clicks on Component 2, then clicks on a formula in the Available Formulas window, and
then clicking on apply again. The user then clicks on Component 3, then clicks on the
formula in the Available Formulas window, and clicks on apply again. After the components
have been selected using this method, the user may click on Components 1, 2, and 3 to view the
formula selected for each component. Once the user has verified that the correct components
are selected, then click OK, and the Vector Plot will be displayed.
Create 3D Mesh Plot uses data for the currently selected formula
at that formula's selected geographic region of interest, layer range,
and time step range, to make a 3D mesh plot.
Create Time Series Line Plot makes a time series line plot
using data for the currently selected formula
at that formula's selected geographic region of interest, layer range,
and time step range. Each time step's
data is averaged linearly to produce that time step's data point.
WARNING: If multiple layers are selected, PAVE 1.4.2 will show data
for only the topmost layer instead of averaging the data over the
selected layers.
Create Time Series Bar Plot makes a time series bar plot
using data for the currently selected formula
at that formula's selected geographic region of interest, layer range,
and time step range. Each time step's
data is averaged linearly to produce that time step's data point.
Create Scatter Plot makes a scatter plot from the formula selected
as component 1 and the formula selected as component 2.
The formulas selected as the components of the vector must match
in geographic region of interest, layer range, and time step range.
- The ScatterPlot popup window is used by the user to specify the formula
for component 1 by clicking on Component 1, then clicking on a formula in
the Available Formulas window, and then clicking on Apply. The user then
clicks on Component 2, then clicks on a formula in the Available Formulas window, and
then clicking on apply again. After the components have been selected using
this method, the user may click on Components 1 and 2 to view the formula selected for
each component. Once the user has verified that the correct components are selected, then
click OK, and the Scatter Plot will be displayed.
Create N-Hour Average Tile Plot creates a tile plot that
contains in the first timestep, the nhour average starting at the
first timestep, in the second timestep, the nhour average starting at
the second timestep, etc.. First, specify the formula using the menu item
Formula, and the menu item Edit/Select from Formula List prior to the
selection of the Create N-Hour Average Tile Plot. The title for tile
plot generated by the Create N-Hour Average Tile Plot menu is
labeled n-hour average, i.e. n-hour average:formula name
Create N-Layer Average Tile Plot creates a tile plot of the
nlayer average for the layers selected using the menu item Formulas,
then the menu Select Layer Ranges Matching Current Formula. The title for tile
plot generated by the Create N-Layer Average Tile Plot menu is
labeled n-layer average, i.e. n-layer average:formula name
Animate Tile Plots Synchronously.. animates all currently
displayed tile plots in a round-robin fashion. As of Version 1.7, PAVE
will synchronize the animation based on the time stamps of the plots,
as opposed to the frame number. As a result, files that start at
different times can be animated synchronously in a meaningful way.
Set Minimum Frame Time.. brings up a window that allows
you to adjust the minimum time between animation frames. This is useful
if you have an extremely fast machine on which you want to slow down the
animation. The minimum time can be set in 0.1 second increments to
any value between 0 and 5 seconds.
Set Tile Plot Cross Section Type brings up a sub-menu that can
be used to choose the axis on which PAVE slices the data to make a plot.
By default, PAVE does a "Z Cross Section" plot, where every data point on
the plot is on the same Z layer. "X Cross Section" and "Y Cross Section"
plot types can also be chosen. Subsequent plots will reflect the chosen
plot type. Choose X Cross Section when you want to plot a cross section
in which the x coordinate is constant, and Y Cross Section when you want
the y coordinate to be constant. Note that you will have to adjust your
geographic region of interest and the layer range appropriately for the
plot type, as PAVE needs to have the geographic region of interest
sufficiently cropped for it to be able to resolve which individual plane
it is plotting. The other sub menu items are currently grayed out, as
PAVE doesn't make use of them. Future versions of PAVE may have these
enabled.
Default slices.. is grayed out as PAVE doesn't use this feature.
The export menu can be used to save data to disk for use by other
programs.
AVS5 saves the currently selected formula's data to an
AVS5 field file, that can then be visualized with the Application
Visualization System (AVS) from
Advanced Visual Systems, Inc.
netCDF saves the currently selected formula's data to a
Models-3 I/O API data file. Data in this format can be visualized later
using PAVE, or any other program that reads netCDF/Models-3 I/O API data
files. You may find it helpful to save your data this way to perform an
extraction from your data, or save the result of a complex formula to
prevent having to recalculate the result.
Tabbed ASCII saves the currently selected formula's data to a
tab delimited data file suitable for reading into a spreadsheet
application such as Excel or Lotus.
Data Explorer and Iris Explorer are grayed out, as this
functionality has not yet been added to PAVE.
The help menu brings up Mosaic loaded with web pages to help you
with your PAVE usage.
User Guide first looks on the local disk for the PAVE
user guide, for <top level PAVE directory>/doc/pave/index.shtml.
If this file does not exist, it attempts to connect to the on-line
user guide installed at CEP, which is at
https://www.cmascenter.org/pave/documentation/2.3/index.html.
Frequently Asked Questions first looks on the local disk for the
PAVE FAQ, for <top level PAVE directory>/doc/pave/Pave.FAQ.html.
If this file does not exist, it attempts to connect to the
FAQ installed on-line at CEP, which is at
https://www.cmascenter.org/pave/documentation/2.3/Pave.FAQ.html.
Models-3 I/O API attempts to use Mosaic to browse the
Models-3 I/O API web pages located at
https://www.cmascenter.org/ioapi/documentation/all_versions/html/AA.html
or https://cjcoats.github.io/ioapi.
Return To Table of Contents
Many aspects of PAVE time series line plots can be configured by the
user. To access the main configure menu, click on the configure button
at the bottom of the time series line plot. Many users like to edit the
minimum and maximum of the axes, and the spacing between and labeling of
the ticks. To configure these attributes, first click on the Axis button
on the configure menu. An Axis Configuration window will appear, and at
the top are buttons that let you choose the axis to edit - select one of
these. To set the maximum and minimum values displayed for the axis,
enter the desired values in the -max and -min fields. To set the
frequency of the tick labels, enter a value for the stepsize. There are
also a number of other features that can be configured.
Many aspects of PAVE tile plots can be configured by the user.
Configuration settings can also be
saved to a file for use in later PAVE plots.
"Control..Configure...Tile..." menu item on a tile plot is used to
adjust the configuration of a tile plot. You will see a window similar to the following:
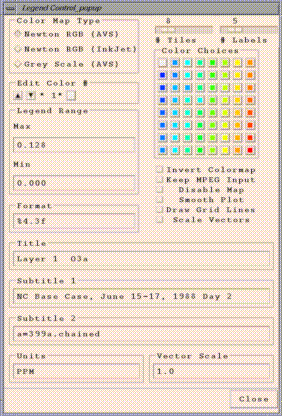
Note that with any of the typein widgets on this window, changes only
take effect when you press the enter key in that widget.
- The Color Map Type radio buttons allow you to choose from three
different color maps.
- The Edit Color # box allows you to individually edit any of the
colors in that tile plot's legend. The arrows enable you to scroll
through the legend's colors until you find the one you wish to change.
You can then modify that color by clicking on one of the Color
Choices color squares to the right. Alternatively, you can modify
that color by clicking on the small color square in the Edit Color # box,
that will then bring up a window that allows you to specify the
individual RGB values for that color.
- The Legend Range box allows you to specify the minimum and maximum
cutoffs for the color legend used by the tile plot. Note that any cells
with values higher than the maximum are plotted with the same color as
the maximum, and cells with values lower than the minimum are plotted
with the same color as the minimum.
- The Format typein specifies a C language sprintf format that is
used to print the labels on the legend. "%4.3f" causes the numbers to print
like "0.128". There are many ways of printing floating point numbers
using sprintf(). Please check your local sprintf() man pages or your
favorite C language text if you need further information on format characters.
- The Invert Colormap toggle reverses the order of the colors in the
given color legend.
- The Keep MPEG Input toggle allows the the user to save the
parameter file and the screen capture (.xwd) files used by mpeg_encode
to create an MPEG animation. When this button is selected, files with the
following names will remain in the same directory as any MPEGs saved from
that tile plot:
[MPEG FILE NAME].param
[MPEG FILE NAME].mpg.0000.xwd
[MPEG FILE NAME].mpg.0001.xwd
:
[MPEG FILE NAME].mpg.N.xwd [where there are N+1 frames in the MPEG]
Note that the MPEG animation will be generated, regardless of
the setting of the "Keep MPEG Input" button.
Parameter files are used by mpeg_encode to generate the MPEG files from a
stream of XWD images saved by PAVE. If you want to edit a parameter file
and then generate an MPEG "manually" using mpeg_encode, the command line
format is:
mpeg_encode <parameter file name>
Using the parameter file below, /home/thorpe/UWINDa2.mpg would
be generated from /home/thorpe/UWINDa2.mpg.[0000-0005].xwd.
IQSCALE 6
PQSCALE 6
BQSCALE 6
PSEARCH_ALG LOGARITHMIC
BSEARCH_ALG CROSS2
GOP_SIZE 10
SLICES_PER_FRAME 1
PIXEL HALF
RANGE 10
PATTERN IBBPBBPBBPBBPBB
FORCE_ENCODE_LAST_FRAME
BASE_FILE_FORMAT PPM
OUTPUT /home/thorpe/UWINDa2.mpg
INPUT_CONVERT /pub/storage/edss/framework/EDSSv0.x/IRIX5_mips/bin/OPTIMIZE/publi
c_domain/xwdtopnm * | /pub/storage/edss/framework/EDSSv0.x/IRIX5_mips/bin/OPTIMIZE/public_
domain/pnmdepth 255
INPUT_DIR /home/thorpe
INPUT
UWINDa2.mpg.*.xwd [0000-0005]
END_INPUT
REFERENCE_FRAME ORIGINAL
FORCE_ENCODE_LAST_FRAME
- The Disable Map toggle allows you to show the data for that plot
without a map.
- The Smooth Plot toggle can be used to do bilinear interpolation on
that tile plot, which provides a smoothed out look at your data. This
slows down the drawing of the plot because PAVE then needs to do
calculations for each pixel of the plot rather than for each grid cell.
- The Draw Grid Lines toggle can be used to turn on grid lines for
your plot, in case this makes it easier for you to see your data's grid
cell size and locations.
- Scale Vectors toggle is only active when the tile plot has vectors on it.
Otherwise, this option is greyed out. By default, the vectors shown are all
the same length. If you want them to be scaled by vector magnitude, turn on this toggle.
If you want to lengthen the vectors, use a scale greater
than one. If you want to shorten them, use a scale less than one.
A future development would be to have a new pull down window to configure
vector tile plots ..Control ..Configure ..ScaleVectors.
- The rest of the items on this window are pretty much self explanatory.
"Control..Configure...Obs..." menu item on the tile plot is used to adjust
the configuration of the Observation data points on a tile plot. This is only active
when the tile plot has vectors on it. Otherwise, this option is greyed out.
You will see a window that will allow you to specify the size and the thickness
of the symbols used for the observational data:

"Control..Configure...Contour..." menu item on the tile plot is used to adjust
the configuration of the Contour plots. This option is only active when the tile plot
has contours on it. Otherwise, this option is greyed out. Need an image showing what this
window looks like
"Control..Configure...VectObs..." menu item on the tile plot is used to adjust
the configuration of the Vectors overlayed with Observational data. This option is only
active when the tile plot has vectors and observational data on it. Otherwise, this option
is greyed out. Need an image showing what this window looks like
"Control..Configure...Title/Subtitle Font..." menu item on the tile plot is used to adjust
the font size of Titles and Subtitles of the tile plot.
Need an image showing what this window looks like
Saving configuration settings for later
use
Tile plot configuration settings can be saved to an ASCII file, edited if
desired, and reused in future PAVE sessions. Using a tile plot's a
"File...Save Configuration Settings" menu item brings up a file browser
that allows you to save the current configuration of the tile plot to an
ASCII file suitable for editing. The information saved includes the
legend range, the format of the legend labels, the number of labels on
the legend, the number of tiles/colors used, the colors themselves, and
all of the individual toggle and radio buttons on the Configure..
window. In subsequent PAVE sessions, these configuration settings can be
retrieved to affect future tile plots.
An example of the file format used is as follows:
ColorMapType NEWTON_COLORMAP
Legend_Max 0.128
Legend_Min 0.000
Legend_Format %4.3f
Number_Labels 5
Invert_Colormap 0
Number_Tiles 8
255 0 0 ColorNumber8
255 147 0 ColorNumber7
218 255 0 ColorNumber6
70 255 0 ColorNumber5
0 255 221 ColorNumber4
0 143 255 ColorNumber3
0 95 255 ColorNumber2
223 223 223 ColorNumber1
Save_MPEG_Files 0
Disable_Map 0
Smooth_Plot 1
Draw_Grid_Lines 1
Scale_Vectors 0
Note the following about the format of the configuration
file:
Configuration files can be loaded into PAVE sessions in two ways -
using the user interface or from the command line. PAVE's main
window now has a "File... Choose Configuration File For New Tile Plots"
menu item, which can be used to select the file. Alternatively, PAVE's
command line arguments or standard input stream can accept input of
the form -configFile <configuration file name>.
Once PAVE loads a configuration file, that information will be
used as the default for all subsequent PAVE plots, until another
configuration file is specified to PAVE.
Return To Table of Contents
8. Printing and exporting images,
animations, and data
PAVE provides facilities to print plots, export images and animations, and export subsets of data. In cases where PAVE's
built in capabilities do not allow you to capture the images you want
(e.g. saving multiple windows together), the tools xv
and snapshot may allow you to accomplish your goals.
You probably need to set your PRINTER environment variable prior to
launching PAVE for printing to work correctly.
(See the Quick PAVE Jump start section for
further information.)
To print tile plots, if you are printing to a black and white printer,
you should probably choose "Control/Configure" from the plot's menu bar and then
experiment with various settings for the colors. You may want to
reduce the number of colors in the plot by choosing a smaller number
of tiles, or alternatively you may want to choose to use the grayscale
colormap. Once you are ready to print the image, just choose the File..Print
menu item on the tile plot, and it will be printed on the printer specified
by the PRINTER environment variable. If for some reason this
doesn't work, then see the sections below on using snapshot
and/or xv. These tools will enable you to save a screen
captured image to a PostScript file that can then be printed.
To print 3D mesh plots, first decide whether you want to print using
black and white or color PostScript. You can then select either the
"Black & White PostScript" or the "Color PostScript" button on the 3D mesh plot's
"ANIMATING SURFACE" window. Then type in the filename, complete with path
name, in the resulting window. Your PostScript will be saved to the chosen
file, and then you can issue an lpr command from the Unix command line to
print that file.
To print time series line plots or scatter plots,
select the PRINT button on the plot's window. Then type in the filename,
complete with path name, in the resulting window. Your PostScript will be
saved to the chosen file, and then you can issue an lpr command from the Unix
command line to print that file.
To print time series bar plots,
see the sections below on using snapshot
and/or xv. These tools will enable you to save an screen
captured image to a PostScript file that can then be printed.
Tile plots can be saved as GIF, Postscript, SGI's RGB, XWD, TIFF,
and several other image file formats using file menu items on the tile
plot.
Tile plots can be saved to an MPEG animation using the
File..Save MPEG Animation menu item on the tile plot.
Several points to note regarding MPEG animations:
- They can take a while to generate.
- They use up a fair amount of disk space, typically 0.4MB per frame
to generate, and 0.2-0.4 MB for the MPEG file for each 24 hours of
animation. This varies
proportionally with the size of the image you are animating. It also
varies according to the number of changes in the image during
animation.
- The MPEG animations can be viewed using mpeg_play, which is
distributed with PAVE as
<pave installation dir> /$EDSS_ENV/bin/OPTIMIZE/public_domain/mpeg_play. Use "mpeg_play -help" for a description of its options.
Here is a useful example:
mpeg_play test.mpeg -loop -framerate 5 -quiet
"-loop" keeps it looping.
"-quiet" keeps it from printing to stdout (which makes the transition from the
last frame to the first much faster).
"-framerate 5" slows it down to print 5 frames per second.
Tile plots can be saved to an animated GIF using the
File..Save Animated GIF menu item on the tile plot.
Several points to note regarding animated GIFs:
- The animated GIF is created by PAVE using an x window dump
- each of the timesteps in the tile plot is then converted to a gif image.
- While the animaged GIF is being created by PAVE, the user must not bring up other windows.
If other windows cover the tile plot while the x window dumps are being performed,
then the animated gif will incorrectly include the contents of these windows rather than
the tile plot window.
- After the x window dumps are completed, a program called convert
creates the animated gif. If there are many timesteps in the dataset, there will be a
delay before the user is again given control of the pave gui. After the convert
program has finished running, control of the PAVE GUI will return to the user.
- The animated GIFs can be viewed using a web browser.
To print other types of PAVE plots,
see the sections below on using snapshot
and/or xv. These tools will enable you to save an screen
captured image to an image file that can then be printed.
PAVE's Export menu enables you to save the currently selected formula's
data in AVS5 field format, netCDF Models-3 I/O API format,
and as tabbed ASCII data suitable for use in spreadsheet
applications. (Note that the currently selected formula's data
is saved when using the export menu items.)
Of particular interest may be the option to export netCDF
data. You might find a very large dataset cumbersome to keep around when
you are only interested in a small number of variables, perhaps in a
small time range or spatial region. PAVE can be used to subselect the
variable, time, levels and region of interest and then save this
data to a smaller, more manageable dataset. Also, you can save the result
of a complex formula to a netCDF file and therefore prevent having to wait
for it to compute again.
On an SGI, type snapshot. To view the snapshot menu, place the
mouse pointer over snapshot and press the right mouse button. Choose the
New file name option to name the file that will be output from
snapshot. You should give it a name with a ".rgb" extension. While the
mouse pointer is over snapshot, press and hold down the shift key. Then
move the mouse pointer to the top left corner of the part of the screen
you wish to capture. Then press the left mouse button and drag the mouse
pointer until you reach the bottom right corner of the section you want
to capture. You should see a red box surrounding the part of the screen
that will be captured. If you need to modify the size or location of the
box, place the mouse pointer over snapshot and press and hold down the
shift button. To move the box, place the mouse pointer inside the box and
then drag the box using the middle mouse button. To resize the box, place
the mouse pointer near one of the edges of the box and drag the edge (or
corner) using the left mouse button.
Once you have selected the portion of the screen to be captured, use
the Save as ... option on the snapshot menu to create an RGB file.
To convert the RGB file to black and white PostScript, type
tops your_file.rgb > your_file.ps
If you want a color PostScript file, type
tops your_file.rgb -rgb > your_file.ps
You can then use xpsview your_file.ps to view the new PostScript
file. You can then capture another image by placing the mouse
pointer over snapshot, pressing and holding the shift button, and
then creating a new box with your left mouse button.
For more information on snapshot and tops, see their man pages.
Sun also has a version of snapshot that can capture images. The images
can be loaded and viewed with imagetool and then saved to a variety of
formats, including postscript. Both snapshot and imagetool have nice
user interfaces to help you through the process. See the man pages
for more information.
xv is a very usfule tool for capturing and manipulating
images - especially GIF files. Type xv to start the program. To view
the xv menu, place the mouse pointer over the inital window and press the
right mouse button. Click on the grab button with the left mouse button,
then use the middle button to drag over the screen region you wish to
save to a PostScript file. A copy of these pixels will appear in a
separate window.
Now left-click the xv controls Save button. Choose
PostScript from the Format menu, and Full
Color or your preferred color choice from the Colors
Menu. Next use the browser to save the PostScript to a file. It is
recommended that you give the file a .ps extension, to indicate its
format.
You can then use xpsview your_file.ps to view the new PostScript
file.
xv is available for a variety of UNIX Platforms via anonymous
ftp to ftp://www.trilon.com/pub/xv
For more information on xv, see the WWW page at
http://www.trilon.com/xv
NOTE: xv is a shareware program that you will need to
pay $25 to license if you decide to use it. This can be remitted to
US Mail: John Bradley
1053 Floyd Terrace
Bryn Mawr, PA 19010
FAX: (610) 520-2042
Electronic Mail regarding XV should be sent to one of these three addresses:
xv@trilon.com - general XV questions
xvbiz@trilon.com - XV licensing & pricing questions
xvtech@trilon.com - bug reports, technical questions
Return To Table of Contents
PAVE has a large number of command line arguments. These can be typed
into the PAVE standard input stream (the window where PAVE was launched),
or supplied via command line arguments when PAVE is invoked. The command
line argument method is often used within scripts written to automate
plots for the user. Notes regarding the scripting commands are available
below. Several example scripts
are provided below. The format for PAVE command line arguments can
be determined by typing "pave -usage" at the command line, which
produces the following:
usage: pave
[-alias <aliasNAME=definition> (NEW in v2.1)!!!]
[-animateWindows<single|continuous> ]
[-animatedGIF<filename> ]
[-autoContourRange]
[-barplotYformat<format string> ]
[-closeWindow<windowid> ]
[-configFile<configFileName> ]
[-contourRange<minCut> <maxCut> ]
[-copyright ]
[-crossSectionType X|Y|Z ]
[-display <display> ]
[ -drawGridLabels ON|OFF (NEW in v2.3!!!) ]
[ -drawLegend ON|OFF (NEW in v2.3!!!) ]
[ -drawMinMax ON|OFF (NEW in v2.3!!!) ]
[ -drawTimeStamp ON|OFF (NEW in v2.3!!!) ]
[ -drawTiles ON|OFF (NEW in v2.3!!!) ]
[-f [<host>:]<filename> ]
[-fulldomain ]
[-g <tile|line|mesh|bar> ]
[-gtype <tile|line|mesh|bar> ]
[-height <tile plot height in pixels> ]
[-help|fullhelp|usage ]
[ -imageMagickArgs 'args' (NEW in v2.3!!!) ]
[-kedamode]
[-legendBins "<bin0,bin1,...,bin_n>" ]
[-level <level> ]
[-levelRange <levelMax> <levelMin> ]
[-mapCounties]
[-mapName "<pathname>/<mapFileName>" ]
[-minMaxModelObs <col> <row> <radius> <formula1> <formula2> ]
[ -minMaxObs <col> <row> <radius> <formula1> <formula2> ]
[ -multivarNcf <formulaList> <varList> <fileName>" ]
[ -multitime <Nformulas> "<formula1>" ... "<formulaN>" ]
[ -nHourAverage <nhours> ]
[ -nHourSum <nhours> ]
[ -nLayerAverage ]
[ -nLayerSum ]
[ -obs <formula> ]
[ -obsidtable <filename> ]
[ -obsSize <size> ]
[ -obsThick<size> ]
[ -obsTimeSeries]
[ -onlyDrawLegend (NEW in v2.3!!!) ]
[ -preClip <llLat> <llLon> <urLat> <urLon> ]
[ -printAlias ]
[ -quit|exit ]
[ -raiseWindow <windowid> ]
[ -s "<formula>" ]
[ -save2ascii <filename> ]
[ -save2d <imagetype> <filename> ]
[ -save2ncf <filename> ]
[ -saveImage "<image type>" <file name> ]
[ -scatter "<formula1>" "<formula2>" ]
[ -showWindow <windowId> <timestep> ]
[ -subDomain <xmin> <ymin> <xmax> <ymax> ]
[ -subTitle1"<sub title 1 string>" ]
[ -subTitle2"<sub title 2 string>" ]
[ -subTitleFont <fontSize> ]
[ -system "<unix command>" ]
[ -tfinal <final time step> ]
[ -tileYlabelsOnRight ]
[ -tinit <initial time step> ]
[ -titleFont <fontSize> ]
[ -titleString "<title string>" ]
[ -ts <time step> ]
[ -tzoffset <Timezone offset> ]
[ -tzset <in Timezone> <out Timezone ]
[ -unalias <aliasname> ]
[ -unitString "<unit string>" ]
[ -vectobs <formula> <formula> ]
[ -vector "<U>" "<V>"]
[ -vectorPlotEvery "<number>"]
[ -vectorScale "<scale factor>"]
[ -vectorTile "<formula>" "<U>" "<V>"]
[ -version ]
[ -width <tile plot width in pixels> ]
[ -windowid ]
-alias <aliasNAME=definition>The user can define an alias by
creating a definition using variable names and derived variables that are calculated using the
mathematical operators described in the PAVE Using Formulas documentation. The alias definition
does not include the dataset name. The alias is treated like any other formula once the alias
definition and the dataset to which it should be applied to is specified. If you need to redefine
an alias definition, you must first use the -unalias command. The alias definitions are saved
to a .pave.alias file in your home directory. Pave uses this type of optional file in your home
directory to maintain a snapshot of the current aliases being used within pave. The following
warning will be reported if an alias is defined more than once: WARNING: Alias <aliasname>
already defined, new definition ignored. The user is also responsible for not making circular
references. If the aliasname is contained within its definition, then PAVE will crash. Use the
-printAlias command to view what aliases are already defined.
An example script to create a tile plot of a user specified alias:
- create a tile plot of the all Nitrogen species
- from the model dataset $DATA_DIR/CHEM_CONC_3D_G1.200021512
#!/bin/csh -f
setenv DATA_DIR /env/data/trayanov
setenv FILE1 CHEM_CONC_3D_G1.200021512
pave \
-f ${DATA_DIR}/${FILE1} \
-alias NTOT=NO+NO2+NO3+N2O5+HONO+HNO3+HNO4 \
-s NTOTa \
-gtype tile \
-unalias NTOT
-animatedGIF<filename>creates an animated GIF by doing an x window dump
of each of the timesteps in the tile plot then converting them to gif images.
While the animaged GIF is being created by PAVE, the user must not bring up other windows.
If other windows cover the tile plot while the x window dumps are being performed,
then the animated gif will incorrectly include these contents of these windows rather than
the tile plot window. After the x window dumps are completed, a program called convert
creates the animated gif. If there are many timesteps in the dataset, there will be a
slight delay before the user is again given control of the pave gui, after the convert
program has finished running.
-autoContourRange tells PAVE to no longer use the contour range
supplied by a previous -contourRange command, but rather use the default
range that is set by the range of the data for each plot.
-barplotYformat"<format string>" can be used to adjust
the format used by sprintf() to draw the y axis labels on bar plots. The default
format is %g. Please see a C reference book or the man pages on sprintf()
for specifics on usable formats for float variables.
NOTE: This option does not work on the IBM platform.
-closeWindow"<windowid>" closes the window with the specified
X window ID
-configFile <configFileName> specifies a configuration
file for PAVE to use for configuring subsequent tile plots. Please
see the Configuring plots section for
more information on configuration files.
-contourRange <minCut> <maxCut> sets the contour
minimum and maximum cutoffs to use for tile plots. By default,
the contour range is set by the range of the data for each plot.
-copyright prints out copyright information on PAVE itself
and all of the third party public domain applications that it uses.
PAVE will not start until the user scrolls throught the copyright information by
hitting rertun or the space key in PAVE's standard input window
-crossSectionType X|Y|Z sets the graphics cross section type
for subsequent plots to the slice type specified.
-drawGridLabels ON|OFF
-drawLegend ON|OFF
-drawMinMax ON|OFF
-drawTimeStamp ON|OFF
-drawTiles ON|OFF were added in version 2.3,
along with other command line arguments that control
whether various parts of tile plots are drawn. You may find this useful
when placing PAVE outputs in other documents. Note that the syntax
to prevent the drawing of titles was already in earlier versions of
PAVE - just give a string of spaces as the argument for the title
command line arguments (e.g. -titleString " " -subTitle 1 " "
-subTitle2 " "). Also see -onlyDrawLegend.
-f [<host>:]<pathname/filename> tells PAVE
to load in this dataset and make it the currently selected dataset.
Each time you enter a new sequence of script commands with -f options in
it, PAVE will remove any previously loaded datasets from its memory,
and begin denoting the new dataset(s) with the letters a, b, etc. So if
you want to load in a number of datasets using the -f option with PAVE's
standard input, be sure to have all the -f <file>
pairs on one line. If you are using a script, it is fine to leave
-f <file> commands on separate lines that end with
backslashes.
-fulldomain sets the PAVE domain matching the currently selected dataset
to be completely selected. The currently selected dataset is usually the
most recently added dataset, unless you have modified it by selecting another
dataset using the user interface.
-gtype <tile|line|mesh|bar> instructs PAVE to create
a plot using the specified type and the currently selected formula's data.
-help | -fullhelp | -usage display the information on all the
command line arguments available. Each of these three versions
perform the identical function.
-imageMagickArgs 'args' was added
in version 2.3.
When used, this command line argument will pass the contents of
args to the convert program that is called when images are converted
from X-window dumps to other image formats. For more information
on the command line arguments for convert, see the ImageMagick
documentation (available from
http://www.imagemagick.org).
-legendBins "<bin0,bin1,...,bin_n>" causes PAVE to use
the specified numbers as breaks between colors on subsequent plots.
The value of this argument is a comma separated list of numbers.
For example, -legendBins &qu;1,10,100,1000&qu; will
cause plots to be created with three colors that correspond to values of
1-10, 10-100, and 100-1000. To go back to the default method for
determining breaks between bins, enter -legendBins DEFAULT.
-level <level> sets the level range of all formulas
to the single level specified.
-levelRange <levelMax> <levelMin> sets the level range of all formulas
to the range specified.
-mapCounties causes PAVE to use the county map for subsequently
created plots. If the PAVE_DISTINCT_STATE_COUNTIES environment variable
is set, the state and county lines will be drawn with different colors.
-mapName "<pathname>/<mapFileName>" causes PAVE
to use the supplied map name instead of the default map for tile plots.
The default map is
<top level PAVE directory>/$EDSS_ENV/bin/OPTIMIZE/maps/OUTLUSAM,
which is a medium resolution state outline map.
-minmaxobs <col> <row> <radius> <formula1> <formula2>
<formula1> = "MODEL_VAR", <formula2> = "OBS_VAR"
creates a plot of the Min and Max of the Model Variable computed over an
area defined with a radius (number of cells) and center cell (Column, Row).
The Observational variable defined at cell (Column, Row) is also displayed
-minmaxmodelobs <col> <row> <radius> <formula1> <formula2>
<formula1> = "MODEL_VAR", <formula2> = "OBS_VAR"
creates a plot of the Min and Max of the Model Variable computed over an
area defined with a radius (number of cells) and center cell (Column, Row).
The Observational variable defined at cell (Column, Row) and the Model Variable
defined at cell (Column, Row) are also displayed
-multivarNcf <formulaList> <varList> <fileName>"export multiple netCDF
variables to a file name. The fileName must currently be listed using the full path name.
An example script to create export multiple formulas to a netCDF file:
- from the model dataset $DATA_DIR/CHEM_CONC_3D_G1.200021512
- create an alias for the total nitrogen species
- load the individual nitrogen species into the formula list
- export the formulas NTOTa, NOa, NO2a to a netCDF file with the
variable names NTOT, NO and NO2, giving the full pathname to the netCDF file
#!/bin/csh -f
setenv DATA_DIR /env/data/trayanov
setenv FILE1 CHEM_CONC_3D_G1.200021512
pave \
-f ${DATA_DIR}/${FILE1} \
-alias NTOT=NO+NO2+NO3+N2O5+HONO+HNO3+HNO4 \
-s NTOTa \
-gtype tile \
-s NOa \
-s NO2a \
-multivarNCF "NTOTa,NOa,NO2a" "NTOT,NO,NO2" $EDSS_ROOT/data/frmwk/pave/multiNTOT.ncf \
-unalias NTOT
-multitime <Nformulas> "<formula1> .. "<formulaN>"
creates a time series line plot showing each of the Nformulas formulas
with its own line.
Note that Nformulas must be between 1 and 8, and that all the formulas for
the plot should have already been loaded into PAVE,
and they are case sensitive.
-NhourAverage <nhours>creates a tile plot that contains in the first timestep,
the nhour average starting at the first timestep, in the second timestep, the nhour average
starting at the second timestep, etc.. The formula specified using -s <formula> prior to the
-NhourAverage option is displayed in the tile plot. The title for tile plot generated
by the -NhourAverage option is labeled n-hour average, i.e. n-hour average:formula name
-NhourSum <nhours>creates a tile plot that contains in the first timestep,
the nhour sum starting at the first timestep, in the second timestep, the nhour sum starting at
the second timestep, etc.. The formula specified using -s <formula> prior to the
-NhourSum option is displayed in the tile plot. The title for
tile plot generated by the -NhourSum option is labeled n-hour sum,
i.e. n-hour sum:formula name
-NlayerAverage <nlayers>creates a tile plot of the nlayer average for the
layers selected using the -levelRange option
-NlayerSum <nlayers>creates a tile plot of the nlayer sum for the
layers selected using the -levelRange option
-obs <formula> plot standard AIRS AMP350 observational
data overlayed on a gridded tile plot with model variable data. The program AIRS2M3
is provided to convert the standard AIRS AMP350 observational data format
to the Models-3 I/O API format that PAVE requires.
-obsidtable <filename> An example lookup table for
converting AIRS AMP350 numeric observation station ids to text names is available
with the airs.lookup file provided with the PAVE 2.1b release. The required format for the
file is a two column, space delimited ascii file, the entries in the first column contain
the AIRS AMP350 numeric observation station ids that can be unquoted or quoted,
the entries for the second column contain the cooresponding name e.g. "CITY, COUNTY, STATE"
that can either be a tightly packed string, or a quoted string. If PAVE finds a match between the
AIRS AMP350 numeric observation station id provided in the gridded observational data file,
and a numeric entry in first column of the airs.lookup file, then it will replace the AIRS AMP350 numeric
observation station id with the cooresponding name provided in the second column of the
airs.lookup file. If no match is found, the AIRS AMP350 numeric observation station id will be preserved
-obsSize <size> specify the size (an integer number)
of the diamond shaped observational data point markers. Specify this option
after the -obs option
-obsThick <size> specify the thickness (an integer number) of the diamond shaped
observational data point markers. Specify this option after the -obs option
-obsTimeSeries creates a time series plot displaying the observations and
average observations versus time. The labels currently list the numeric observation id.
To focus in on specific counties or regions use the -subdomain option.
An example script application:
- create a tile plot of the model Ozone data overlayed with observations
- the observations are plotted as diamonds, and shaded with a line thickness of 3, and the size of 6
- create a timeseries plot of observation data and average observation data
- for the subdomain region (26,72) (3,50)
- from the observation dataset $DATA_DIR/O38.ids.obs_on
- from the model dataset $DATA_DIR/SM_b1on_cc3_g0.l1.O3.8hr.2hours
#!/bin/csh -f
setenv DATA_DIR $EDSS_ROOT/data/frmwk/pave/
setenv FILE1 O38.ids.obs_on
setenv FILE2 SM_b1on_cc3_g0.l1.O3.8hr.2hours
pave \
-f ${DATA_DIR}/${FILE1} -s O38_OBSa \
-f ${DATA_DIR}/${FILE2} -s O3b \
-subdomain 26 3 72 50 \
-gtype tile -obs O38_OBSa \
-obsSize 6
-obsThick 3
-obstimeseries
-onlyDrawLegend ON|OFF
was added in version 2.3.
If this is set to ON, you'll get a plot for which only the
legend is drawn. This is useful if you want to place the legend
in another document. You can crop this plot when exporting images
using scripts by using the -imageMagickArgs command line option.
-preClip <llLat> <llLon> <urLat>
<urLon>
will cause PAVE to use a "pre-clip" map region bounded by the
supplied lat/lon coordinates. These arguments can be
used to access PAVE's world map over a region outside of North
America.
For further information on how to use the world map, please see the
PAVE FAQ located at
Pave.FAQ.html#WorldMap.
-printAlias prints existing alias definitions
-quit | -exit ends the PAVE session.
-raiseWindow "<windowid>" raises the window with the specified
X window ID (i.e. brings it to the front)
-s "<formula>" loads the specified formula into PAVE's memory,
and makes it the currently selected formula.
-save2d <imagetype> <filename> used to save a timeseries plot to
an image file. Supported imagetypes include GIF, RGB, XWD, MPEG, or PS. The environment variable
GRAPH2DCFG is used to specify the graph2d.blt BLT configuration file. An example graph2d.blt BLT
configuration file is provided with the PAVE 2.1b release.
-save2ascii <filename>export data to a tab delimited data file suitable for
reading into a spreadsheet application such as Excel or Lotus
-save2ncf <filename>export data to a Models-3 I/O API netCDF formatted file.
Data in this format can be visualized later using PAVE, or any other program that reads
Models-3 I/O API netcdf data files.
-saveImage <image type> <file name>
saves the most recently created tile plot,
The image type can be RGB, XWD, GIF, MPEG, or PS.
Note: this may cause PAVE to crash if the most recently created tile plot has
been closed.
This option can also be used to generate a stream of images from a tile
plot, one for each time step of data associated with that
tile plot. This can be done either through a command line
option (-saveImage) or through the standard Motif user
interface that comes with a tile plot's window. By
supplying a file name with % format characters suitable by
the C Language's printf() routine, PAVE is notified
that a stream of images should be saved rather than a single
image. The printf % format characters are used to generate
the individual file name for each time step of data.
For example, if a user needs to:
- save Gif images of variable UWIND for the first 13 hours
- from dataset /home/thorpe/example_data/399test.wind.bin.d1
- to files /tmp/test00.gif .. /tmp/test12.gif
- with smoothly colored background (as opposed to tiled)
- with a contour range of -4000 to +4000
- from a script, quitting PAVE upon completion.
The following script would do the job:
#!/bin/csh
setenv SMOOTH_PLOTS 1
pave \
-f /home/thorpe/example_data/399test.wind.bin.d1 \
-s UWINDa \
-contourRange -4000 4000 \
-tinit 0 -tfinal 12 -gtype tile -saveImage GIF /tmp/test%02d.gif \
-quit
-scatter "<formula1>" "<formula2>" creates a scatter
plot using the two formulas specified. Note that the formulas for the
two components should have already been loaded into PAVE, and they are case
sensitive.
-showWindow <windowId> <timestep> Sets the time step
of the window with the specified X-window ID to the specified timestep.
The timestep must be within the allowable range for the dataset.
-subdomain <xmin> <ymin> <xmax> <ymax>
sets the PAVE domain matching the currently selected dataset to the
bounding box specified by its arguments. The currently selected dataset
is usually the most recently added dataset, unless you have modified it
by selecting another dataset using the user interface. It is often handy
to type -subdomain commands into PAVE's standard input if you are trying
to select a very precise subdomain (such as that needed for a vertical
cross-section plot).
-subTitle1 and -subTitle2 allow
the user to control a tile or vector plot's subtitles if desired.
Subsequent plots will use the default subtitles, unless these
arguments are used again.
-subTitleFont"<fontSize>"allow the user to control
the font size of the subtitle of a pave plot.
-system "<unix command>" sends the specified command to the
UNIX command line using C language's system() routine.
-tfinal <final time step> sets the last time step
for each formula's time step range to the specified step number, where the first
step number is denoted by 0.
-tileYlabelsOnRight causes the tile plot's Y axis labels to appear
on the right hand side of the plot, rather than the default of the
left hand side.
-tinit <initial time step> sets the first time step
for each formula's time step range to the specified step number, where the first
step number is denoted by 0.
-titleFont"<fontSize>"allow the user to control
the font size of the title of a pave plot.
-titleString "<title string>" sets the title for the
next plot made to the specified title. Subsequent plots will use
the default PAVE title, unless this argument is used again.
-ts <time step> sets the selected time step for each
formula in PAVE's memory to the specified step number, where the first
step number is denoted by 0.
-tzoffset <Timezone offset>changes the time by the specified offset
value, use a +/- integer value. This option is specified after the -gtype tile option
-tzset <in Timezone> <out Timezone> The <out Timezone> will be
displayed in parenthesis next to the time on the tile plot. The correct specification
of the <in Timezone> can not be verified by PAVE. It is the responsibility
of the user that the timezone specified as <in Timezone> matches the native timezone
of the input data.
Currently Supported Timezones, (use the acronym in all caps)
GMT, Greenwich Mean Time; CET, Central European Time; EET, Eastern European Time
AST, Atlantic Standard Time; EST, Eastern Standard Time; CST, Central Standard Time
MST, Mountain Standard Time; PST, Pacific Standard Time; YST, Yukon Standard Time
HST, Hawaii Standard Time
-width and -height
allow the user to control a tile plot's width and height in pixels.
All subsequent plots will use the supplied value for height/width, until
a non-positive value is supplied as a subsequent argument. At that point
PAVE's default height/width will be used.
-unalias <aliasname>used to undefine an alias
-unitString can be used to override the default unit label
used for tile plots. The default value comes from the dataset(s)
themselves.
-vector "<U>" "<V>" creates
a vector plot with U as the
left to right vector component and V as the down to up vector component.
There are no background colors used for this type of plot.
Note that the formulas for the two components should have already
been loaded into PAVE, and they are case sensitive.
-vectobs <formula> <formula> plot wind vector observational data overlayed
on a vector tile plot of model wind vector data.
An example script application:
- create a vector tile plot of the model wind vector data overlayed with wind vector observations
- color the wind vector observational data blue, to distinguish from the model wind vector
obs that are drawn in black
- change the scale of the wind vectors to reduce the wind vector length
- from the observation dataset $DATA_DIR/TDL_OBS.avg.smraq
- from the model dataset $DATA_DIR/mm5_winds.smraq.lyr_1.avg.full
#!/bin/csh -f
setenv DATA_DIR1 $EDSS_ROOT/data/frmwk/pave/
setenv FILE1 mm5_winds.smraq.lyr_1.avg.full
setenv FILE2 TDL_OBS.avg.smraq
setenv PAVE_VECTOBS_COLOR blue
pave \
-f ${DATA_DIR1}/${FILE1} \
-s U_AVGa -gtype tile\
-s V_AVGa \
-f ${DATA_DIR1}/${FILE2} \
-s U_OBSb \
-s V_OBSb \
-vectorScale 15 \
-vectortile U_AVGa U_AVGa V_AVGa \
-vectobs U_OBSb V_OBSb
-vectorPlotEvery <number> causes PAVE to plot every n vectors
where n is equal to the number specifed (n must be an integer).
-vectorScale <scale factor> uses scale (an integer number) to
adjust the length of non-uniform vectors. This needs to be specified before the
-vectortile option
-vectorTile "<formula>" "<U>" "<V>" creates
a vector plot with the result of "formula" as the background tiles, U as the
left to right vector component, and V as the down to up vector component.
Note that the formulas for the three components should have already
been loaded into PAVE, and they are case sensitive.
-version prints out information about the PAVE version
being used on the standard output stream, that is usually
sent to the window PAVE was launched in.
-windowidPrint the X-window ID of the most recently created window.
Here are a few notes to keep in mind regarding the above scripting
commands:
- default host will be local
- default formula will be most recently added
- default level will be level 1
- default time step will be 0 (0 based)
- default initial time step will be 0 (0 based)
- default final time step will be Nsteps in file-1 (0 based)
- host can be expressed as either a file name or an ip number
- files must be either UAM, UAM-V, or netCDF format
- datasets (filenames) will be tagged (by the PAVE subsystem) "a", "b", ...,
in the order PAVE learns of them
- default domain will be all on.
Here is an example PAVE script:
compare.pave:
#!/bin/csh -f
setenv DATA_DIR /ep/otag/jul93/output
setenv SCEN1 07bas1B
setenv SCEN2 93snsDuc2
pave \
-f $DATA_DIR/$SCEN1/bin/avrg.cc.22jul93-07.mc.$SCEN1 \
-f $DATA_DIR/$SCEN2/bin/avrg.cc.22jul93-93.mc.$SCEN2 \
-configFile /ep/otag/jul93/output/base_o3.cfg \
-unitString "PPM" \
-titleString "$SCEN1 Ozone" \
-s O3a -tinit 12 -tfinal 17 -gtype tile \
-titleString "$SCEN2 Max Ozone" \
-s max\(O3b\) -tinit 12 -tfinal 18 -gtype tile \
-configFile /ep/otag/jul93/output/compare_o3.cfg \
-titleString "$SCEN1 Ozone - $SCEN2 Ozone" \
-s O3a-O3b -gtype tile
This script compares ozone concentrations in coarse grid average files
for two scenarios and draws three plots. In the first few lines some
environment variables are set to help find data in a generic fashion.
pave \
-f $DATA_DIR/$SCEN1/bin/avrg.cc.22jul93-07.mc.$SCEN1 \
-f $DATA_DIR/$SCEN2/bin/avrg.cc.22jul93-93.mc.$SCEN2 \
Here pave is started and two files are loaded (coarse grid average files
for a day of interest).
-configFile /ep/otag/jul93/output/base_o3.cfg \
In the next line a configuration file is defined. This contains preferences
for "base ozone plots" (i.e. plots of ozone concentrations, not differences).
The contents of the file are:
base_o3.cfg:
ColorMapType NEW in v2.1TON_COLORMAP
Legend_Max 0.160
Legend_Min 0.000
Legend_Format %4.3f
Number_Labels 7
Invert_Colormap 0
Number_Tiles 8
Save_MPEG_Files 0
Disable_Map 0
Smooth_Plot 0
Draw_Grid_Lines 0
Scale_Vectors 0
Here the standard colormap is being used, max and min values are set, and the
number of labels is changed to 7. The rest of the values are defaults - they
do not need to be in the configuration file. You can create a configuration
file by configuring a plot as you want it and choosing Save Configuration
Settings from the file menu of the plot.
-unitString "PPM" \
-titleString "$SCEN1 Ozone" \
-s O3a -tinit 12 -tfinal 17 -gtype tile \
In these lines, setup is being done up to plot ozone for the first data
set. The units and title are defined, the formula to plot is chosen, and
in this case only hours 12-17 will be plotted. Finally, a tile plot is
created due to the keywords "-gtype tile". NOTE: the \'s at the ends of
the lines are continuation characters - don't forget these when you're
using PAVE from a C-shell script!
-titleString "$SCEN2 Max Ozone" \
-s max\(O3b\) -tinit 12 -tfinal 18 -gtype tile \
In these lines a plot of the maximum ozone in dataset b is created by
using the max operator. The \'s around the ()s are required because
C-shell will try to interpret the ()s otherwise. Note that by using
tinit and tfinal, the maximum is for those hours only, but the resulting
file will contain multiple time steps with the same value for each one.
-configFile /ep/otag/jul93/output/compare_o3.cfg \
-titleString "$SCEN1 Ozone - $SCEN2 Ozone" \
-s O3a-O3b -gtype tile
In the last three lines a difference plot is drawn with different preferences
as defined by a new configuration file. The rest is similar to the other
plots. Here is the configuration file for the difference plot:
compare_o3.cfg:
ColorMapType JET_COLORMAP
Legend_Max 0.032
Legend_Min -0.032
Legend_Format %4.3f
Number_Labels 9
Invert_Colormap 0
Number_Tiles 9
Huge example of command line usage:
Here is a huge example of invoking PAVE with command line arguments. Almost
all available command line arguments are used by this example. These
types of invocations typically are used from within a script that can
be run over and over. Of particular note are the vectorTile,
vector, scatter, and multitime options, that
are only available through command line
or via stdin (but NOT via the PAVE user interface).
pave \
-f /pub/storage/edss/framework/EDSSv0.2/example_data/399a.uam.emis.d1 \
-f /200p_scratch/olerud/399a/uamdata/399a.wind.bin.d1 \
-s "sqrt(UWINDb*UWINDb+VWINDb*VWINDb)" \
-s UWINDb \
-contourRange -3.0 3.0 \
-titleString "Ground Level U Wind Speed" \
-gtype tile \
-s VWINDb \
-contourRange -3.0 3.0 \
-titleString "Ground Level V Wind Speed" \
-gtype tile \
-autoContourRange \
-titleString "Ground Level Winds (Bckgnd=Speed)" \
-vectorTile "sqrt(UWINDb*UWINDb+VWINDb*VWINDb)" "UWINDb" "VWINDb" \
-vector "UWINDb" "VWINDb" \
-s "NO2a+NOa" \
-titleString "Ground Level NOx Emissions" \
-gtype tile \
-titleString "Ground Level NOx Emissions" \
-gtype mesh \
-titleString "Ground Level NOx Emissions" \
-gtype line \
-titleString "Ground Level NOx Emissions" \
-gtype bar \
-scatter "UWINDb" "VWINDb" \
-f /200p_scratch/rmm/eastUS_em1_g0 \
-s "NOc+NO2c" \
-contourRange 0 200 \
-subdomain 26 24 37 34 \
-level 1 \
-titleString "Ground Level NOx" \
-gtype tile \
-level 5 \
-titleString "Level 5 NOx" \
-gtype tile \
-levelRange 1 5 \
-contourRange 0 200 \
-subdomain 31 24 31 34 \
-crossSectionType X \
-titleString "First 15 Levels NOx at Col 31" \
-gtype tile \
-contourRange 0 200 \
-subdomain 26 30 37 30 \
-crossSectionType Y \
-titleString "First 15 Levels NOx at Row 30" \
-gtype tile \
-s "NOc" \
-s "NO2c" \
-multitime 2 NOc NO2c
Return To Table of Contents
The following environment variable settings can be made prior to
invoking PAVE. They are completely optional, and most of them
can also be set from within PAVE's user interface.
You may find these useful if you frequently want smooth plots,
grid lines, no maps, etc.
setenv PAVE_EXE <some pathname/executable name>
will cause the pave script to use that version of the
binary executable when launching PAVE.
setenv PAVE_DEBUG <ON|OFF> in interpreted by the
pave script in versions 2.3 or later. If PAVE_DEBUG is set to ON
(default is OFF),
then the script prints some debugging information about the various
paths used when PAVE is executed. This is useful if you are having
problems getting PAVE or one of its components to work. You can
turn this on by doing setenv PAVE_DEBUG ON before running PAVE.
setenv KILL_PROCESS_GROUP <ON|OFF> is interpreted
by the pave script in version 2.3 or later.
If KILL_PROCESS_GROUP is set to ON (which is the
default behavior in the pave script), then all processes started by
PAVE during the session (e.g. time series plots) will be killed when
PAVE exits. However, we have found that on Linux, if PAVE is being
run from a script the parent script is also killed. Therefore, you
may wish to do setenv KILL_PROCESS_GROUP OFF before running PAVE
on Linux - especially if you are using scripts.
setenv LOCAL_FILES_ONLY <ON|OFF> is interpreted
by the pave script in versions 2.3 or later. If LOCAL_FILES_ONLY is
set to ON (the default value
is OFF), PAVE will execute without running the software bus. This is
useful when running from scripts because you can run multiple instances
of PAVE from the same user account. A limitation is that when
LOCAL_FILES_ONLY is ON and you are running PAVE in interactive mode,
we have been unable to get the local file browser to communicate
back to PAVE properly when new files are added. However, files that
were added in prior sessions or via the command line may be analyzed.
setenv DENOMINATOR_CUTOFF <some floating point number>
will cause PAVE to workaround divide by zero conditions.
If this environment variable is set, then for each
division, if the denominator is less than or equal to
DENOMINATOR_CUTOFF's value, then the result of the divide is
set to 0
setenv DISABLE_MAPS 1
will cause PAVE to disable map drawing by default on tile plots
setenv DRAW_GRID_LINES 1
will cause grid lines to be drawn by default on tile plots
setenv ELLISPOID ellipsoidName will cause PAVE to use a
non-default ellipsoid when drawing maps. The valid values for
ellipsoidName are:
MERIT_1983, SGS_85, GRS_1980, IAU_1976, CLARKE_1866, CLARKE_1880
ENGELIS_1985, EVEREST_1969, WGS_60, WGS_66, WGS_72, and WGS_84.
See also the entries on this page for SPHERICAL_EARTH and GRS80.
setenv EDSS_MAPDIR <directory_name>
will override PAVE's <top level PAVE directory>/$EDSS_ENV/bin/OPTIMIZE/maps dir,
which is where PAVE looks for its maps by default.
setenv GRS80 will cause PAVE to assume the earth's ellipsoid
is the one defined by the GRS80 specifications. The defalt ellipsoid
is MERIT_1983. See also SPHERICAL_EARTH and ELLIPSOID on this page.
setenv LEGEND_BINS &qu;1,10,100,1000&qu; for example will
cause plots to be created with three colors that correspond to values of
1-10, 10-100, and 100-1000. The general format for the value of this
environment variable is a comma separated list of numbers.
setenv NO_AUTHOR_STRING will cause PAVE not to put PAVE by MCNC on
plots.
setenv PAVE_EXPORT_VARNAME <some alphanumeric string>
can be used to override the default variable name of "VAR" when exporting
data to netCDF files from PAVE.
setenv PAVE_DISTINCT_STATE_COUNTIES will cause PAVE to display
state and county lines in different colors (gray / black).
setenv PAVE_VECTOBS_COLOR <colorName> will cause PAVE to draw the
observational vectors with the specified color. Potential colors and RGB values are
usually found in /usr/lib/X11/rgb.txt for non-Suns or /usr/openwin/lib/X11/rgb.txt
on Suns.
setenv PRECLIP_LLLAT <0>
setenv PRECLIP_LLLON <-180>
setenv PRECLIP_URLAT <85>
setenv PRECLIP_URLON <70> will cause PAVE
to use a "pre-clip" map region bounded by the lat/lon coordinate
pairs (-180,0) and (70,85). These environment variables can be
used to access PAVE's world map over a region outside of North
America.
For further information on how to use the world map, please see the
PAVE FAQ located at
Pave.FAQ.html#WorldMap.
setenv PRINTER 200p_lw
will cause tile plots to be sent to 200p_lw when printed
setenv SBUS_EXEC_RC <rc file name> can be used to
override the default file ~/.edss_exec_rc, which is used
to locate various EDSS subsystems
setenv SBUS_RLOGIN_RC <rc file name> can be used to
override the default file ~/.edss_rlogin_rc, which is used for
the userids to use for remote data access. You will only need
to worry about this if your remote account's login name is not
the same as your local login name.
setenv SCALE_VECTORS 1
will cause vector length to be scaled by their magnitude by default
on vector plots
setenv SMOOTH_PLOTS 1
will cause PAVE to smoothly interpolate pixels on tile plots by default
setenv TENTHS_SECS_BETWEEN_FRAMES 10
will cause a minimum of 1 wall clock second between animation frames.
Units are tenths of a second, allowable range is 0..50
setenv PAVE_COORD "GDTYP P_ALP P_BET P_GAM XCENT YCENT"
<some real number> can be used to define the projection
parameters. CAMx or UAM files in the Lambert conformal projection are
not self-contained, as the lambert parameters are not specified in the file header.
To inform PAVE that the CAMx or UAM file has the conformal projection,
specify the values using the PAVE_COORD environment variable.
GDTYP,P_ALP,P_BET,P_GAM,XCENT,YCENT are defined in the the IOAPI documentation
http://www.baronams.com/products/ioapi/GRIDS.html
An example for a lambert conformal grid with a reference lat/lon of (40N,100W)
and latitudes of two parallels, 60N and 30N. Note XCENT is typically equal to
P_GAM.
setenv PAVE_COORD "2 30 60 -100 -100 40"
setenv [ P_ALP|P_BET|P_GAM|XCENT|YCENT|XORIG|YORIG|XCELL|YCELL ]
<some real number> can be used to supersede any of the respective
netCDF header values. Before
creating a PAVE plot, the environment variables will be checked and
appropriate fields replaced.
setenv SPHERICAL_EARTH radius will cause PAVE to treat the earth
as a sphere with the specified radius when drawing maps. If you set the
value of radius to 1, it will use the default radius of 6370997 meters.
Note that the default ellipsoid is MERIT_1983. See also the entries
on this page for GRS80 and ELLIPSOID.
setenv UAM_MISSING_VALUE <some floating point number>
can be used to define a value that corresponds to missing data for
UAM format files. If this environment variable is not set, the value
defaults to -999.0.
setenv USE_LOCAL_VISD will cause PAVE to read all files on the local
computer with the visd instead of as local files. This is useful if a file
to be visualized on a 64 bit capable SGI is larger than 2 GB. This should
only be used at times when very large files are being visualized because
it is slower than reading files locally.
setenv VECTOR_COLOR <colorName> will cause PAVE to draw the
vectors with the specified color. Potential colors and RGB values are usually
found in /usr/lib/X11/rgb.txt for non-Suns or /usr/openwin/lib/X11/rgb.txt
on Suns.
setenv ADD2DHLINE <some floating point number>
used to define a horizontal line to a 2-D plot such as a timeseries plot.
setenv GRAPH2DCFG <blt filename> This specifies a BLT configuration file
that can be modified to configure 2D plots. All 2D plotting in PAVE is generated using
the BLT library, which is an extension to the (Tcl)/Tk library.
setenv HOST <machine name or ip address> used to specify a remote hostname
from which to obtain a file, used in the -f option.
Return To Table of Contents
You must have access to either a
Sun running Solaris 2.x, an SGI running IRIX 5.x, a DEC Alpha
running OSF1, an IBM RS6000 running AIX 4.x, an HP running
HP-UX 9.x, a PC running Linux, or a PC running Windows NT/XP/2000 with
Interix (aka Microsoft Services for Unix) installed. Note that PAVE will
also work on higher versions of most of the operating systems specified here.
You must have your input datasets in Models-3 I/O API (netCDF)
UAM-IV, or UAM-V formatted data files.
PAVE can be displayed on most X displays with at least 8 bits of color.
Using X windows software on a PC or Macintosh, you should
be able to to display PAVE output from any of the above Unix
platforms.
(OPTIONAL - READ THE REST OF THIS SECTION THIS ONLY IF YOU WANT TO ACCESS REMOTE DATA)
PAVE uses several optional files in your home directory
when starting up, and writes over them with each PAVE session
termination. These are used to maintain a snapshot of the current
formulas, datasets, and aliases being used within PAVE:
~/.edss_rlogin_rc # you may need to create if using remote data
~/.edss_exec_rc # PAVE will create if not already there
~/.pave_history_rc # PAVE will set this up automatically
~/.pave.alias # PAVE will set this up automatically
~/.pave.AA.cases # PAVE will set this up automatically
~/.pave.AA.formula # PAVE will set this up automatically
The only one that really matters is ~/.edss_rlogin_rc, which
is used for setting up remote data accessibility. Here are the contents
of an example ~/.edss_rlogin_rc file, which you can borrow
from to create your own. Also note the instructions for setting up
remote .rhosts files and paths to the visd and busd daemons.
# ~/.edss_rlogin_rc
#
# -------------------------------------------
# If you want to use PAVE to read remote data:
# -------------------------------------------
# This file will need to be used by PAVE to launch a
# daemon on any remote machine(s). The daemon(s) actually
# read the data and ship it back to your local machine.
#
# Copy this file to *your* ~/.edss_rlogin_rc and modify
# it appropriately
#
# On each machine you will need to set up a ~/.rhosts
# file that allows THIS machine to rsh to it. On most machines
# a .rhosts file is a list of machine and login name pairs
# found in your home directory (e.g. sirrocco smith). Make sure
# that this file is readable only by you for security reasons.
#
# Suppose you are running PAVE on sirocco, and you want to use it
# to read data that is sitting on sequoia. Test to see that your
# .rhosts file on sequoia works by making sure the following
# commands on sirocco:
#
# rsh sequoia.nesc.epa.gov -l <YOUR sequoia USERID> which visd
# rsh sequoia.nesc.epa.gov -l <YOUR sequoia USERID> which busd
#
# successfully execute and tell you the paths to the visd and busd
# daemons on the remote sirocco machine. The visd and busd daemons
# are in <pave installation dir>/bin/
# for each platform type; on remote machines you may just wish to
# copy them to your home directory if it makes it easier for you.
# These daemons are used to read remote data; until they are in your
# remote path, you won't be able to read any data on that remote
# machine.
#
# Any line in this file with a # in it is ignored.
#
####################################################################
# $HOME/.edss_rlogin_rc file contains the userids for remote machine
# names. If remote userid is same as local, you don't need to list it.
# Lines preceded by a '#' are ignored.
# The format of the contents in the file are:
# <machine-name><space><userid>
# The machine name can be the entire name or without the domain name
# (eg. nox, rain)
####################################################################
#mary.jane.doe doe
sequoia.nesc.epa.gov tsr
t90.ncsc.org demo_t90
The other files will be set up for you automatically
whenever PAVE executes.
Return To Table of Contents
This section can be used to guide you through an example PAVE session.
If you are a new user, it can help you become familiar with most
of PAVE's features. Once you have satisfied the
Requirements for use section above, then:
- Login to the machine where PAVE is installed. If it is a remote
machine, setenv your
DISPLAY environment
variable to wherever you are sitting, as in
setenv DISPLAY mymachine.wherever.gov:0
If you will be printing tile plots directly from PAVE, you may
also want to set your PRINTER environment variable appropriately.
For example, if you
setenv PRINTER 200p_qms
then PAVE will use print-command lpr -P200p_qms to print
tile plots. Otherwise PAVE will use the print-command
lpr, which defaults to the printer lp, and
may or may not be accessible to your system.
- Launch PAVE from the command line by typing pave at the Unix
prompt. You will probably need to make sure the scripts subdirectory
of the top level PAVE installation directory into your path in order
for your shell to be able to resolve the pave location. The
AAREADME file that comes with PAVE has information on how
to do this.
Typing "pave" launches a "wrapper script", which in turn
fires up the software bus and the PAVE executable itself. The PAVE
executable itself should never be launched directly from the command
line, as this wrapper script is required to set up PAVE's
environment.
- You should see an "Add/Delete/Select Dataset" pop-up window:
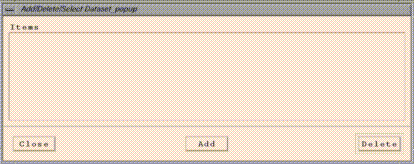
Click on its "Add" button, and an EDSS file browser should appear:
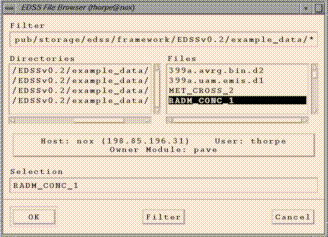
(this may take a
couple seconds). The file browser has a "Filter" widget at the top. You
can see the files in a particular directory by typing or pasting in the
directory name and adding a "/*" at the end and hitting return.
Alternatively, you can navigate through the directory structure by double
clicking on directories in the Directories list (or single clicking and
hitting return). Once you get to the directory that contains the file
you want, you can select the file by clicking on it in the Files list.
Use the file browser to add the file
"<top level PAVE directory>/example_data/RADM_CONC_1"
(If RADM_CONC_1 is not available, choose any dataset in the
directory.) This will become PAVE's dataset "a", and this will be the
currently selected dataset. Do the same with
<top level PAVE directory>/example_data/399a.uam.emis.d1
which will become dataset b.
There may be several other example datasets in <top level PAVE
directory>/example_data that you may want to play around with,
although some may be missing due to disk space considerations:
-rw-r--r-- 1 thorpe edss 8549788 Aug 17 09:26 399a.avrg.bin.d2
-rw-r--r-- 1 thorpe edss 1070068 Aug 17 09:27 399a.uam.emis.d1
-rw-r--r-- 1 thorpe edss 11870588 Aug 17 09:29 MET_CROSS_2
-rw-r--r-- 1 thorpe edss 11870584 Aug 17 09:30 RADM_CONC_1
-rw-rw-r-- 1 thorpe edss 7871036 Aug 17 09:32 bot_west_chem_ec1_g0
-rw-r--r-- 1 thorpe edss 201048 Aug 17 09:32 exact1
- After adding the dataset, you should see a "Species List" window:
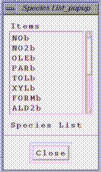
This should now be displaying the species available in dataset b, the
currently selected dataset.
- Go back to the "Add/Delete/Select Dataset" window
and "Select" the first dataset (denoted by "a") by clicking on it.
This now becomes the currently selected dataset,
and you should now see that dataset's available variables in the
"Species List" popup. Click on "HOa" to add that variable to
the available formulas list.
- You should see an "Add/Delete/Select Formula" window:
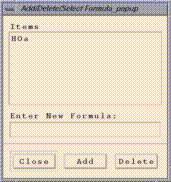
Add item "HOa+HO2a" in the "Enter New Formula:" typein box
and click on "Add". Add "NOb" and "O3a" as well.
You now have a total of four formulas added to your
formula list, and O3a is the currently selected formula.
- When PAVE window is up, select the following sequences from
the menu bar at the top of PAVE window:
"Graphics/Create Tile Plot"
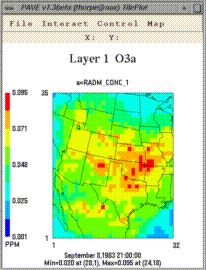 "Graphics/Create 3d Mesh Plot"
"Graphics/Create 3d Mesh Plot"
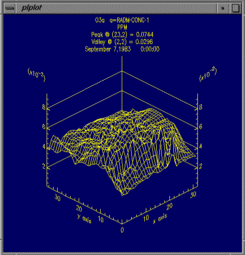 "Graphics/Create Time Series Line Plot"
"Graphics/Create Time Series Line Plot"
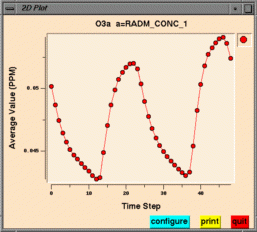 "Graphics/Create Time Series Bar Plot"
"Graphics/Create Time Series Bar Plot"
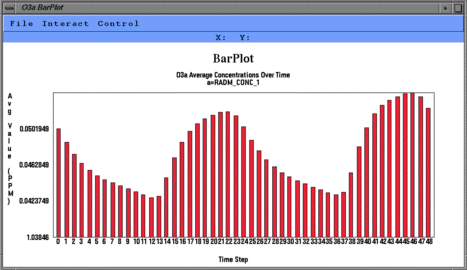
NOTE: All the plots above are associated with the selected
formula, which is probably O3a. The selected formula
can be modified with the "Formulas menu/ Edit/Select from Formula List"
window.
The tile plot and mesh plot show data for the selected region at a
particular time step, and can be animated to show other time steps. The
time series line and bar plots show data averaged over the selected
region at each time step. Here are some things to try with the plots:
- If you want to adjust a tile plot's legend min/max cutoffs,
choose "Control/Configure" from the plot's menu bar.
- If you want to animate a tile plot over time, select
the "Control/Animate" menu item on a tile plot. This
brings up a window from which you can start the
animation.
- If you want to animate all existing tile plots synchronously,
select the "Graphics/Animate Tile Plots Synchronously" menu
item from the main PAVE window. This presents a window
that you can use to start the animations.
- When using a slider (such as in the Animate window for Tile Plots), to
scroll by one time step in either direction, click on the slider
itself and then use the left and right arrow buttons on the
keyboard.
- Select "Formulas/Select Region of Interest Matching Current Formula"
from the menu bar. The domain window associated with the selected formula
will be displayed.
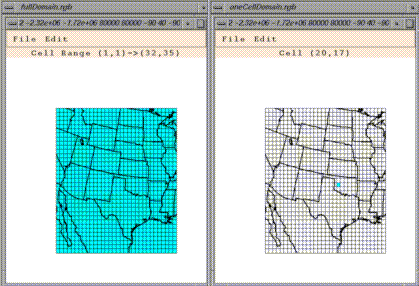
By default, the entire domain is selected for any dataset/formula
loaded into PAVE. This is indicated by a light blue background on
all of the cells in the domain window. You may want to do a time
series of a single cell, or perhaps a tile plot of a smaller region
of cells. To subselect a region, just left-click-and-drag over
any blue areas to deselect cells, and conversely over
any white areas to select cells. Also, you can use
the Edit menu's "Select All" and "Select None" items to
turn all the cells in the domain on or off.
Subsequent plots will show data only from the subselected region.
This window can be closed using the File menu's "Close" item.
Also note the File menu's "Save Domain To File" and
"Load Domain From File" menu items that allow you to
save and later retrieve frequently used domain subselections.
- How to add a remote dataset:
If you have followed the optional instructions under
point 3. of the Requirements to use PAVE section of
this document, you can add remote datasets using the
file browser provided to choose datasets.
Select "Datasets/Edit/Select from Dataset List" from the menu bar. An
Add/Delete/Select Dataset window will appear. Click on the
"Add" button, and an EDSS file browser should appear (this may take a
couple seconds).
Now click on the large button in the center of the file browser that
contains "Host:", "User:", and "Owner Module:" information. This
brings up a "host selector" window in which you can enter the remote
host name (e.g. sequoia.nesc.epa.gov). Clicking on Select should
then enable you to browse for files on the remote machine, starting
from your home directory.
- NOTE: If you want to do a vector plot, a scatter plot of two
formulas, or a time series line plot with multiple (2-8) variables
plotted on it, then you must send commands to PAVE via stdin or
command line arguments, which are described in the
"Driving PAVE using scripts" section of this document.
Unfortunately there are currently no graphical user interface
methods to access these features.
There are examples of invoking
PAVE with command line arguments in the same section.
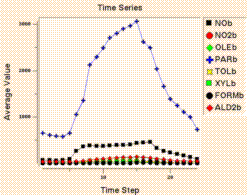
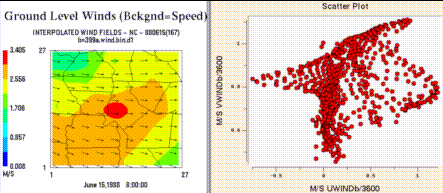
- Explore the menu items available to you from tile plot windows.
- The background map can be toggled between county boundaries,
medium (the default map) or high resolution state boundaries,
rivers, or roads.
- Tile plots can be printed, or saved as
PostScript, XWD, RGB, or Gif images.
- You can zoom in on, probe the data values in, and generate time
series plots for specific regions of tile plots.
- To view a vertical cross section of data using the Tile Plot, first
click on the dataset for which you would like to view the cross
section. Then under the datasets menu, choose "Select Layer Ranges Matching
Current Dataset". You should see a window with sliders for the highest and
lowest layers to use. Set these appropriately depending on which layers you
want to see. To see a vertical cross section, the lowest and highest should
be different layers (as opposed to just plotting a horizontal cross section -
in that case the min and max layers are set to the same value). [Note that
you can also select a layer range for a formula using the corresponding
options under the Formulas menu.]
After selecting the layers you want to see, type a statement of the
following form into PAVE's standard input:
-subdomain 80 20 80 90
Here the arguments to -subdomain are: x1 y1 x2 y2 to define the region
to be selected. This will cause the data where x = 80, and y is between
20 and 90 to be selected. To see this region on the screen (and/or
to set this region using the User Interface rather than a command line
option), you can choose "Select Regions of Interest Matching Current Dataset".
A window showing the domain will come up.
Alternatively, you can select the cells with the mouse, but this
gets difficult when grid cells are small and you want to select a very
precise area.
Now that the layers and region to be plotted are selected, you need to set
the cross section type. Under the Graphics menu, choose "Set Tile Plot
Cross Section Type" and you will see a submenu of X, Y, or Z cross sections.
To make a plot for the above example choose "X Cross Section" (remember this
by noting that x is constant). Next, draw the plot for a formula using
that dataset by choosing "Create Tile
Plot" from the Graphics menu, and you will see the plot of that cross section.
- Explore the rest of the menu items not documented above &
enjoy!
Return To Table of Contents
The UNC Institute for the Environment BugZilla has been
out of service for quite some time. For bugs and comments, please
use https://github.com/cjcoats/pave-3.0
- In versions prior to PAVE 2.1, the PAVE formula parser was
not smart enough to determine the difference between
plotting TA for dataset n (TAn) and using the tan
function. Similar problems might occur with the
combination CO and s. Workaround: make your dataset
use a different letter.
- If the file .pave_exe.pave_usage.log
is not writable by the user, versions prior to 1.5 would
print out some information and then crash with a
segmentation fault or a memory fault. PAVE
1.5 gracefully exits and gives a meaningful error message.
- In versions prior to PAVE 2.1, Mesh plots didn't display
on some non-8 bit graphics displays. This problem has
been solved by using convert from ImageMagick. For PAVE
1.7 and below the problem occurred on non-8 bit graphic
displays (e.g. a DEC 5000 X-Server, some SGIs, Linux
machines, some Windows machines). Workaround (for
versions prior to PAVE 2.1.0): set your display to use
8 bit graphics. On Windows machines this can be done by
configuring your display Color Palette to use 256 colors.
On Linux, the X-server can be started with an 8 bit depth
using startx -bpp 8.
- Probing in vector plots causes PAVE to crash in
version 1.7 and earlier. This is fixed in version 1.7.1.
- On Linux, "Close" buttons do not work properly for
some windows.
Workaround: close the windows using the window manager
decorations.
- Time series plots over a region do not work if all of
the data in the selected region is missing for one of the
selected time steps.
- PAVE sometimes prints a message like
Can't allocate ramp color 214
and runs out of colors to use.
Workaround: exit
PAVE, exit out of any other applications running on your
X-display that are using lots of colors (AVS,
drawing/painting programs, etc), and then get back into
PAVE. If you were running netscape, try
restarting it with the option -ncols 64 to limit
the number of colors it uses.
- Occasionally when saving tile plot images to data files, the
menus "disappear" from the tile plot.
They are still active however they are
"invisible".
Workaround: iconify the tile plot, de-iconify it,
and you should be able to see the menus again.
- Printing tile plots may not work reliably on the Sun platform
at this time, for reasons unknown to us.
Workaround: save your images as Gif or RGB, convert
them to PostScript (see the Saving PAVE
Plots to PostScript section for more information on
this), then print them.
- The non-default maps in PAVE are a bit slow to generate,
especially the first time you create a plot with a given
map projection. We hope to speed up the map generation
process with future versions of PAVE.
- If you choose "File/Exit" and PAVE does not shut down,
there may be a mesh plot or time series line plot still
running that prevents PAVE from exiting. Workaround:
locate any line or mesh plot windows that you have
created, and quit out of them before exiting PAVE.
- Miscellaneous memory leaks.
Workaround: If you run PAVE for a large number of
plots and you notice performance degradation, exit out of
PAVE and then restart it.
- In PAVE 1.4.x, PAVE could export netCDF files,
but had trouble reading them back in if the exported
formula was at all complex. This is corrected in version
1.5. See New Features in PAVE
1.5 for more details.
- In versions prior to PAVE 1.5, the column numbers in
tabbed ASCII files output from PAVE were sometimes
incorrect. This has been corrected in PAVE 1.5.
- In versions prior to PAVE 1.5,
if you use the tile plot Control..Time Series menu
item to generate a time series plot from a tile plot that
was generated by first selecting a subdomain, PAVE
may not show the correct data.
Workaround: (1) Download the latest PAVE;
(2) Use this menu item only from plots that are not on a
subdomain - zoom in on the subdomain instead and then
generate the plot; (3) select the subdomain for which you
want the time series plot and choose Create Time Series
Line Plot from the Graphics menu to create the plot.
- In versions prior to PAVE 1.5,
creating a time series plot from the Graphics menu
when there are multiple layers of data selected
generates a plot for the top data layer only, instead of
for the data averaged over the selected layers. Version
1.5 reports an error message if the user requests a time
series plot over multiple layers.
- In versions prior to PAVE 1.5, there was a bug reading
UAM-IV wind files. This has been corrected in
PAVE 1.5.
Return To Table of Contents
Malloc failure: If PAVE prints or pops up a message of the
form malloc failure or VIS_DATA_Dup()
failed this means that PAVE has run out of usable memory
while trying to perform the requested operation. To make more
memory available, try closing some plots or quit and restart PAVE.
Can't allocate ramp color xxx: If PAVE prints a message about
not being able to allocate a color that means that all available colors
for the current session on your system have been used up. Applications
like netscape will use as many colors as possible. To keep more colors
available to applications like PAVE, try running netscape with the option
-ncols 64 to prevent it from getting all the colors. This problem may
also occur if you've selected a lot of different color schemes during
your current PAVE session. To get all colors in the desired color scheme
try quitting and restarting PAVE after you've chosen the scheme that you
like. Also, logging out of your current session and logging back in
should make the maximum number of colors available.
Return To Table of Contents
The pave script now responds to the environment variable
KILL_PROCESS_GROUP. If KILL_PROCESS_GROUP is set to ON (which is the
default behavior in the pave script), then all processes started by
PAVE during the session (e.g. time series plots) will be killed when
PAVE exits. However, we have found that on Linux, if PAVE is being
run from a script the parent script is also killed. Therefore, you
may wish to do setenv KILL_PROCESS_GROUP OFF before running PAVE
on Linux - especially if you are using scripts.
The pave script now reponds to the environment variable
LOCAL_FILES_ONLY. If LOCAL_FILES_ONLY is set to ON (the default value
is OFF), PAVE will execute without running the software bus. This is
useful when running from scripts because you can run multiple instances
of PAVE from the same user account. A limitation is that when
LOCAL_FILES_ONLY is ON and you are running PAVE in interactive mode,
we have been unable to get the local file browser to communicate
back to PAVE properly when new files are added. However, files that
were added in prior sessions or via the command line may be analyzed.
The pave script now reponds to the environment variable
PAVE_DEBUG. If PAVE_DEBUG is set to ON (default is OFF),
then the script prints some debugging information about the various
paths used when PAVE is executed. This is useful if you are having
problems getting PAVE or one of its components to work. You can
turn this on by doing setenv PAVE_DEBUG ON before running PAVE.
- A set of new command line arguments was added that control
whether various parts of tile plots are drawn. You may find this useful
when placing PAVE outputs in other documents. Note that the syntax
to prevent the drawing of titles was already in earlier versions of
PAVE - just give a string of spaces as the argument for the title
command line arguments (e.g. -titleString " " -subTitle 1 " "
-subTitle2 " "). The new commands to control whether the other
parts of the plot are drawn are:
-drawLegend ON|OFF
-drawMinMax ON|OFF
-drawTimeStamp ON|OFF
-drawTiles ON|OFF
-drawGridLabels ON|OFF
A new command line argument -onlyDrawLegend ON|OFF
was added. In this case, you'll get a plot for which only the
legend is drawn. This is useful if you want to place the legend
in another document. You can crop this plot when exporting images
using scripts by using the -imageMagickArgs command line option.
The command line argument -imageMagickArgs 'args' was added.
When used, this command line argument will pass the contents of
args to the convert program that is called when images are converted
from X-window dumps to other image formats. For more information
on the command line arguments for convert, see the ImageMagick
documentation (available from
http://www.imagemagick.org).
PAVE now responds to the environment variable ELLIPSOID
to specify the earth's ellipsoid. See Environment
Variables page for more information.
You can now specify the radius of the earth and treat the earth
as a spher using the environment variable SPHERICAL_EARTH.
See Environment
Variables page for more information.
Updated COPYRIGHT info to GNU Public License.
Removed references to MCNC.
PAVE by MCNC is no longer put on plots by default.
The map region is no longer preclipped by default.
Added PNG as a format to which images can be exported.
PAVE now kills BLT and other child processes upon exit. This works for Interix, but should be tested more thoroughly on other platforms.
The limitation on the maximum number of timesteps was removed. Now annual hourly files can be analyzed.
Updated Makefiles to have more utility.
Corrected problem with very large and small numbers changing to scientific notation then causing problems in formulas.
-
The numerical divisions between the color bins to be controlled by the
user. In earlier versions of PAVE, the divisions between the color
bins were calculated automatically based of the specified minimum value,
maximum value and the number of bins used. The range of each bin
was the same. For example, each bin might be 10 PPB of ozone, or 1 m/s.
Users can now control the bin divisions by setting the environment variable
LEGEND_BINS or the command line argument legendBins to a comma separated
list that contains the divisions between the bins. The advantage of the
command line argument is that you can change the values of the bins during
the PAVE session.
After using the legendBins command line argument, you can set the bin
determination to use the default method by entering -legendBins
DEFAULT.
The following example causes a plot to be drawn with three
colors with values in the ranges 1-10, 10-100, and 100-1000:
setenv LEGEND_BINS "1,10,100,1000"
- On SGI 64 bit capable computers, files larger than two gigabytes
can now be displayed. This is done using a 64 bit capable "visd"
on the SGI, which then transfers the selected data to the main PAVE
interface. The only restriction is that amount of selected data must be
less than 2 Gbytes. If the main PAVE program is being executed on the
SGI that holds the large data files, PAVE can be forced to use the visd
to read data files if the user sets the environment variable USE_LOCAL_VISD.
This should not be set unless large files are being visualized
because it is slower than reading a local file directly. If the large
file is being browsed remotely and resides on a 64 bit capable SGI, the
64 bit capable visd will be started by default (if it is installed).
Examples of 64 bit capable SGIs are Octanes, O200s, and O2000s. Some O2s
are 64 bit capable, and some are not.
- County and state lines can be displayed in different colors (gray /
black) if the environment variable PAVE_DISTINCT_STATE_COUNTIES is set.
This will occur when the user selects "Counties (SLOW the first time)"
from the Tile window plot's Map menu.
The different color state and county lines can be obtained when
using a script by issuing the -mapCounties command line argument and
setting PAVE_DISTINCT_STATE_COUNTIES. Note that after the two color county
map has been displayed, it may not be possible to switch to another map
during the same PAVE session.
- Vector plots and vector tile plots have a "vector legend", which
will be turned on if the user selects "Scale Vectors" from the Configure
window. The value of the Vector Scale type-in from the same window
represents the length of the vector legend and the rest of the vectors are
scaled accordingly. This makes the Vector Scale more meaningful than
in previous versions, although the behavior is somewhat opposite (i.e. the
larger the Vector Scale, the smaller the vectors because the legend is
always the same size).
- Vectors can be drawn with a user specified color. This option
is controlled by setting the VECTOR_COLOR environment variable to any
color name recognized by the X-Windows system. Potential colors and
RGB values are usually found in /usr/lib/X11/rgb.txt for
non-Suns or /usr/openwin/lib/X11/rgb.txt on Suns.
- Handling of missing data and time steps other than 1 hour for
I/O API files is improved.
Data in I/O API files with values less than or equal to
AMISS3 (-9.0E36), such as BADVAL3 are now treated as missing data.
This means that no color is assigned on tile plots and values are treated
internally as the IEEE constant for "Not a number". Also, it is now
possible to export netCDF files with time steps other than 1 hour, and
a more current version of the I/O API is used.
- Animations are now synchronized based on their times, not their
time steps. If Animate Tile Plots Synchronously was chosen in
previous versions of PAVE, the first time step of each file was shown
followed by the second, etc. In the current version, the system
examines the actual time the data corresponds to an animates the plots
synchronously.
- Problems with saving multiple screen captures from a script
using %02d have been corrected. In prior versions, there was a
problem when the user tried to save GIF files for all time steps of a
dataset from a script using the syntax
-tinit 0 -tfinal 12 -gtype tile -saveImage GIF /tmp/test%02d.gif
- Corrections were made to visualizing some UAM-IV and UAM-V files.
In earlier versions, UAM-IV boundary files were not visualized properly
unless the number of grid cells in the X & Y directions were the same.
Also, version 1.7.1 supports the visualization for pressure in UAM-V height/
pressure files, and cwater in UAM-V cloud/cwater files whereas in earlier
versions only the height and cloud variables were available, respectively.
- Commands entered from standard input now support single and double
quotes, thus allowing strings such as titles with multiple words to
be passed directly to PAVE from standard input.
- Window management with command line arguments is improved. The
-windowId command line argument now returns the X-window ID of the most
recently created window. The time step displayed by a window can be
controlled using the -showwindow windowId timestep command line
argument. A window can be closed with the -closeWindow windowId
command line argument. A window can be raised (brought to the front)
with -raiseWindow windowID.
- The maximum number of timesteps for a dataset has been increased
such that a datasets with up to 120 days of hourly data can be visualized.
- The Linux version no longer gives errors about "XmDeleteIemsPos",
and is statically linked so external libraries are not needed.
- The Solaris file browser can now display remote files.
- The rare cases when map lines would extend below the visualization
area or black boxes would appear over the date/time information have
been fixed, and a bug that occurred when map lines had more than 2500
vertices has been corrected.
- It is possible to hide the "PAVE by MCNC" string on PAVE plots (e.g.
for publication purposes).
- Up-to-date I/O API and netCDF libraries were used. This corrected
a problem with reading the data from netCDF files on HP-UX.
- PAVE no longer leaves Browser processes running after it is exited.
- When you zoom in to a region, the region plotted always shows a whole
number of grid cells (before, partial cells could be plotted and probing
at the edge of the plot caused an error message).
- DEC OSF version is compiled with native C and C++ so that missing data
(represented as NaN - Not a Number) is treated correctly
- The configure window was made smaller so that it can be displayed on a
1024 x 768 PC monitor.
- Vector plots now display all vectors by default.
- A slider has been placed on the Configure window to control how
frequently vectors are plotted
- Command line options were added: -vectorScale real# controls the
scale of the vectors, -vectorPlotEvery integer (=N) plots every N vectors.
- A bug in time series plots has been corrected.
- IRIX 6.x is now supported by the PAVE script.
- A script pavewin that starts PAVE in its own window is now included.
- A bug that caused pave to crash when time series plots were attempted on formulas
with division has been corrected.
- CONNECTION TO BUS HAS CRASHED message no longer appears when PAVE is
exited normally.
- The user guide has been split into multiple files for faster access
from the web.
- I/O API / netCDF boundary files can now be visualized.
- Missing data is now supported in UAM and netCDF files. Areas with
missing data are shown in the background color of white. For UAM files,
the default value for missing data is -999.0. This can be overridden
with the environment variable UAM_MISSING_VALUE.
- The numeric format specified in the configure window for a tile plot
is now also used to format the numbers in the tabular display when probing
the data.
- Dates with the format YYMMDD are now supported for UAM-IV format files.
- The limit for the length of a formula has been increased to 2500
characters.
There are several bug fixes with this release:
- If you generate a time series plot from the Interact menu for a plot
of a subdomain, the correct data is now displayed.
- NetCDF files produced by PAVE using Export to netCDF can
now be read back in to PAVE. The default species name
for the output variable is
VAR. This can be
overridden by setting the environment variable
PAVE_EXPORT_VARNAME to another alphanumeric
string. Information about the formula that was exported
and the datasets used to create it is stored in the
FDESC portion of the netCDF file. This can be
viewed using the ncdump utility.
- A bug in the reader for UAM-IV winds has been corrected.
- The column numbers in tabbed ASCII output files has been corrected,
and the time stamp labels are now all written on their own lines.
- If the file .pave_exe.pave_usage.log
is not writable by the user, the system now gracefully
exits and gives a meaningful error message instead of
producing a segmentation fault.
- If multiple layers are selected and a time series plot is requested,
an error message saying that this operation is not supported now appears.
This may be supported at some point in the future.
There are several minor bug fixes with this release:
- A bug was fixed with the world maps which prevented domains that
crossed the greenwich meridian from displaying a map.
- A bug was fixed that prevented netCDF "chained" files from
being read into PAVE.
- Integer data in netCDF can now successfully be read by PAVE.
PAVE now can display a world map for data which falls outside
the North America region. For further information on how to use
the world map, please see the PAVE FAQ located at
Pave.FAQ.html#WorldMap.
- The following operators have been added to PAVE's formula parser:
Op Name Function
-- ---- --------
a < b Less than Returns 1 if a < b, else 0
a <= b Less than or equal Returns 1 if a <= b, else 0
a > b Greater than Returns 1 if a > b, else 0
a >= b Greater than or equal Returns 1 if a >= b, else 0
a == b Equal Returns 1 if a equals b, else 0
a != b Not equal Returns 1 if a does not equal b, else 0
a && b And Returns 1 if a != 0 and b != 0, else 0
a || b Or Returns 1 if a != 0 or b != 0, else 0
Please note that PAVE's operator precedence (highest to lowest)
is as follows. If you wish to override the precedence given below,
or are uncertain as to which operator will take precedence, you can
feel free to use parentheses in your formulas. This will force
expressions within the parentheses to be evaluated first.
Highest 1) abs, log, sqr, sqrt, exp, ln, sin, cos, tan, sind, cosd,
Precedence tand, minx, miny, minz, max, maxy, maxz, mean, min, max,
sum, mint, maxt
2) **
3) /, *
4) +, -
5) <, <=, > >=
6) ==, !=
7) &&
Lowest 8) ||
Precedence
- Variables from UAM and netCDF formatted data files can
now be mixed within the same formula, and a suitable map
will be produced, provided the variables lie on the same domain
and have the same map projection. In addition, minor
discrepancies in the precision of the map information
associated with the variables will not prevent PAVE from
drawing a map in the plots.
- PAVE now "plots" formulas that result in a single number
(for example,
mean(O3a)) by sending that single
number to the text window, along with information about the
time and domain currently selected.
- The data is now labeled much more clearly on all time series plots.
The annotations include information on the spatial and
temporal subdomin used to get the data for the plot, as
well as the datasets used.
- There is now a black frame drawn around a tile plot's color legend.
This makes the legend much more readable when white is chosen
as one of the colors.
- PAVE now displays "PAVE by MCNC" in tiny letters on
tile plots and bar plots.
- There are now several new command line arguments
that can be given to PAVE. They are:
-barplotYformat <format string>
-
<format string> is a format character
string that will be used by sprintf() to
draw the y axis labels on bar plots. The default
format is %g. NOTE: This option does not
work on the IBM platform.
-tileYlabelsOnRight
- The
-tileYlabelsOnRight causes the tile plot's
Y axis labels to appear on the right hand side of the
plot, rather than the default of the left hand side.
-subTitle1 "<sub title 1 string>"
-subTitle2 "<sub title 2 string>"
- The
-subTitle1 and -subTitle2 arguments allow
the user to control a tile or vector plot's subtitles if desired.
-height <tile plot height in pixels>
-width <tile plot width in pixels>
- The
-width and -height command line
arguments have been added to PAVE, to allow the user to
control a tile plot's width and height in pixels. All
subsequent plots will use the supplied value for
height/width, until a non-positive value is supplied as
a subsequent argument. At that point PAVE's default
height/width will be used.
-copyright
- The
-copyright option prints out copyright
information on PAVE itself and the third party
"external library" applications that it uses.
-system "<System Command>"
- The
-system option allows the user to supply a
command to be fed to the Unix command line using a call
to the system() library routine.
- There are several user interface changes in this version.
- The "close" and "quit" menu items
have been removed from the Motif menu accessed from
the upper left hand corner of a tile plot window or a
time series bar plot window.
- A Tile Plots "Configure..." window has been
reorganized to be smaller, in order to fit on smaller
displays.
- Several other minor user interface improvements
have been made.
- There are several bug fixes in this version.
- A bug with colors and tile plot configuration files
has been resolved. Users can now select a
configuration file with color information in it, make
plots, and then select a configuration file without color
information, and the original colormaps will correctly be
used.
- Closing a tile plot while it is animating no longer
causes PAVE to crash .
- Chained files with greater than one layer no longer
cause PAVE to crash.
- Double clicking on the upper left hand corner of the
main PAVE window, a tile plot window, or time series bar
plot window will now have no effect . Previously,
this caused PAVE to immediately crash.
-
NaN (bogus not-a-number) data in a
tile plot no longer causes PAVE to crash.
Instead, a warning message is printed to the
stderr stream, and no color is plotted in
that part of the image.
- PAVE no longer kills all processes in its process
group when exiting. This means that parent processes
(such as VisDriver or the EDSS Console) will remain alive
when PAVE exits. However it also means that any mesh
plots will prevent PAVE's completion, until they are quit
out of individually.
- Several other minor bugs have been fixed.
Version 1.4 beta fixes an X-Window related bug that caused
PAVE to crash occasionally with certain selected domains of interest.
]
T
here are a number of new features in this version that you may find
useful, including:
-
There is now a Help menu in PAVE, from which the user
can bring up the PAVE User Guide, the PAVE FAQ, and the
M3 I/O API Web pages using Mosaic.
-
PAVE makes better use of memory. Now, if you close a window, PAVE
releases all memory associated with that window. Thus, if you see error
messages like "malloc failed" or "VIS_DATA_Dup failed" you can try closing
windows to free up memory. Deleting formulas and datasets will also cause
some memory to be freed (and reduce PAVE's start up time). In addition
to freeing memory upon closing windows, PAVE uses memory more efficiently
in general.
-
PAVE now allows the user to edit individual colors used
in tile plots, using a Color Chooser window that
enables setting of the individual red, green, and blue
components on a 0..255 scale using sliders. This window is
accessible from a tile plot's "Control...Configure.."
window, by clicking on the color shown in the
"Edit Color #" box. For ideas on colors to use, see the file
/usr/lib/X11/rgb.txt
for non-Suns or
/usr/openwin/lib/X11/rgb.txt
on Suns.
-
Vector plots now work with vertical cross sections.
-
Vector plots with no tile backgrounds no longer show a color legend.
-
There are now two additional titles that are user controllable
on the PAVE tile plots. These can be changed using a tile plot's
"Control...Configure.." window. They are called
"Subtitle 1" and "Subtitle 2". Subtitle 1 is usually initialized
to an empty string (and therefore not shown on the plot), but if
a UAM data file is used to create the plot, then the file_id
character string will be used. Subtitle 2 is initialized to a list
of datasets used to make that plot.
-
There is now a vector scale that is user controllable.
Using a vector plot's "Control...Configure.." window, the
user can select the "Scale Vectors" radio button to scale
them by vector magnitude, as opposed to the default which is
to use a unit vector length for all vectors. Once "Scale Vectors"
is selected, then the value contained in the "Vector Scale"
typein widget will be used to scale the length. The default
vector scale is 1.
-
A Tile Plot's File menu now has the following menu items:
"Time Series At Max Point" and "Time Series At Min Point",
which find the location of the min or max value in that
tile plot's data, and plots a time series of the data at
that point.
-
A Tile Plot's Interact menu now has a "Time Series" menu
item. This interact mode causes a time series to be
generated for a tile plot's data over a point or region
that is dragged or clicked on.
-
The PAVE formula parser now accepts formulas to calculate
the change in a variable per time step, using the syntax
d[<variable name>]/dt.
For example, d[O3a]/dt would calculate the change in concentration
of ozone from dataset A per time step. Given dataset A with N timesteps,
this formula will actually give you only N-1 timesteps. This
is because each hour I's calculation is computed using
O3a:I+1 - O3a:I
Note that only "atomic" variables may be used in formulas
using this operator. You cannot make such formulas any
more complex than the above example, ie the following
would be ILLEGAL: d[O3a+O3a]/dt and d[O3a]/dt+O3a.
-
PAVE's min and max operators have been modified to calculate
the minimum/maximum value over the selected time period
at each cell. For example, if the user has the formula
max(O3a),
then each cell's data for that formula would be the maximum
value of ozone occurring during the time period currently
selected.
-
While MPEG frames are being generated and written
to disk, the user does not have control of PAVE. However,
MPEG encoding is now done by a background process, in order
to return PAVE's control to the user as soon as possible.
When the MPEG file has been created, the user will be notified
by a beep from their terminal.
-
If the user wishes to save the parameter file used by mpeg_encode
to create an MPEG animation, there is an option under a tile
plot's Control...Configure window to enable this. Checking
on the "Keep MPEG Input" button will cause the following files
to remain in the same directory as any MPEGs saved from that
tile plot:
[MPEG FILE NAME].param
[MPEG FILE NAME].mpg.0000.xwd
[MPEG FILE NAME].mpg.0001.xwd
:
[MPEG FILE NAME].mpg.N.xwd [where there are N+1 frames in the MPEG]
Note that the MPEG animation will be generated regardless of
the setting of the "Keep MPEG Input" button.
Parameter files are used by mpeg_encode to generate the MPEG
files from a stream of XWD images saved by PAVE. If you want
to generate an MPEG file "manually" using mpeg_encode, the
command line format is:
mpeg_encode <parameter file name>
For the parameter file below, /home/thorpe/UWINDa2.mpg would
be generated from /home/thorpe/UWINDa2.mpg.[0000-0005].xwd.
IQSCALE 6
PQSCALE 6
BQSCALE 6
PSEARCH_ALG LOGARITHMIC
BSEARCH_ALG CROSS2
GOP_SIZE 10
SLICES_PER_FRAME 1
PIXEL HALF
RANGE 10
PATTERN IBBPBBPBBPBBPBB
FORCE_ENCODE_LAST_FRAME
BASE_FILE_FORMAT PPM
OUTPUT /home/thorpe/UWINDa2.mpg
INPUT_CONVERT /pub/storage/edss/framework/EDSSv0.x/IRIX5_mips/bin/OPTIMIZE/public_domain/xwdtopnm * | /pub/storage/edss/framework/EDSSv0.x/IRIX5_mips/bin/OPTIMIZE/public_domain/pnmdepth 255
INPUT_DIR /home/thorpe
INPUT
UWINDa2.mpg.*.xwd [0000-0005]
END_INPUT
REFERENCE_FRAME ORIGINAL
FORCE_ENCODE_LAST_FRAME
-
Tile plot configuration settings can now be saved to an ASCII
file, edited if desired, and reused in future PAVE sessions.
PAVE tile plots now have a "File...Save Configuration Settings"
menu item that brings up a browser to save the current
configuration of the tile plot to an ASCII file suitable for
editing and modification by the end user. Information saved
includes the colors, number of tiles, number of labels,
legend range, legend format, and all of the individual
radio buttons on the Configure.. window. In subsequent
PAVE sessions, these configuration settings can be retrieved
to affect future tile plots.
An example of the file format used is as follows:
ColorMapType NEWTON_COLORMAP
Legend_Max 0.128
Legend_Min 0.000
Legend_Format %4.3f
Number_Labels 5
Invert_Colormap 0
Number_Tiles 8
255 0 0 ColorNumber8
255 147 0 ColorNumber7
218 255 0 ColorNumber6
70 255 0 ColorNumber5
0 255 221 ColorNumber4
0 143 255 ColorNumber3
0 95 255 ColorNumber2
223 223 223 ColorNumber1
Save_MPEG_Files 0
Disable_Map 0
Smooth_Plot 1
Draw_Grid_Lines 1
Scale_Vectors 0
Note the following about the format of the configuration
file:
Configuration files can be loaded into PAVE sessions in two ways -
using the user interface or from the command line. PAVE's main
window now has a "File... Choose Configuration File For New Tile Plots"
menu item that can be used to select the file. Alternatively, PAVE's
command line arguments or standard input stream can accept input of
the form -configFile <configuration file name>.
Once PAVE loads a configuration file, that information will be
used as the default for all subsequent PAVE plots, until another
configuration file is specified to PAVE.
-
The "Formulas...Edit/Select From Formula List" window has
been modified somewhat to facilitate easier additions of a number of
formulas. From left to right, the buttons on now read
"Add", "Close", and "Delete". The first
formula typed in by a user should be followed by a click on the
"Add" button. Once that occurs, the default button will be
"Add". Each subsequent formula need only be followed by a
carriage return, which will then perform the same function as clicking
on the "Add" button.
-
PAVE now can generate a stream of images from a tile
plot, one for each time step of data associated with that
tile plot. This can be done either through a command line
option (-saveImage) or through the standard Motif user
interface that comes with a tile plot's window. By
supplying a file name with % format characters suitable by
the C Language's printf() routine, PAVE is notified
that a stream of images should be saved rather than a single
image. The printf % format characters are used to generate
the individual file name for each time step of data.
For example, if a user needs to do the following:
1) save Gif images
2) for the first 13 hours
3) saved to files /tmp/test00.gif .. /tmp/test12.gif
3) of variable UWIND
4) from dataset /home/thorpe/example_data/399test.wind.bin.d1
5) with smoothly colored background (as opposed to tiled)
6) with a contour range of -4000 to +4000
7) from a script, quitting PAVE upon completion
The following script would do the job:
#!/bin/csh
setenv SMOOTH_PLOTS 1
pave \
-f /home/thorpe/example_data/399test.wind.bin.d1 \
-s UWINDa \
-contourRange -4000 4000 \
-tinit 0 -tfinal 12 -gtype tile -saveImage GIF /tmp/test%02d.gif \
-quit
-
Several small changes have been made to make
the user interface easier to use. For example, reselecting a menu item
that causes a window to appear will bring the window to the front, even if it
is buried.
-
Several bugs that caused crashes have been fixed.
There are a number of new features in this version that you may find
useful, including:
- User controllable unit labels
- The default color map for tile plots has been
improved
- MPEG animations can now be saved under Solaris 2.5
- The IBM version has been upgraded from AIX3 to AIX4
- Several bugs that caused crashes have been fixed
- Saving RGB, PostScript, XWD, and Gif images
- Saving MPEG animations
- On-the-fly map projections, which enables any
netCDF dataset conforming to the M3 I/O API standard
to be handled by PAVE
- Several maps can be toggled between, including
low and high resolution state boundaries, rivers,
roads, and county boundaries.
- Reading datasets that are spread across multiple
files
- Exportation of netCDF Models-3 I/O API data files
- Saving/retrieval of selected geographic domains
- Select All/Select None features in the region of
interest selection boxes
- Printing directly from a PAVE tile plot
- Multiple time series (1-8) can now be overlaid
on the same plot
- The min/max for the displayed data is shown on tile plots
- Command line arguments for many of the above features
Return To Table of Contents
PAVE is no longer supported by CMAS; its distribution is now
currently done by Carlie J. Coats, Jr., on an un-funded personal
basis, for the good of the environmental modeling community and
because it is more accessible than many alternatives, and can be
adapted to support huge data sets (unlike some of the
alternatives).
PAVE is now widely used by the Air Quality Modeling Community. We
are appreciative of the communities contributions and adoption of
this visualization product. We are seeking to pool together
funding to support requests for enhancements and bug fixes from
multiple groups to enable the continued support of this product.
If you or someone you know is interested in some of these features
and if you are able to provide funding, or ideas about how to
obtain funding to continue development, please contact me via
email carlie@jyarborough.com.
Some of the features CMAS would have liked to add, as of version 2.3,
are listed below. If you know of additional features that the community
would benefit, please contact me as described above.
- Support CF-format netCDF
See http://cfconventions.org/
- Update external software
It would be a good idea to update PAVE to use the most recent versions of
BLT for time series plots, Tcl/Tk, and PLPLOT.
Done, at least partially, for pave-3.0.
- Update MPEG generation to use ImageMagick
Currently the MPEG generation uses an old technique. It would be good
to update this to use ImageMagick to generate MPEGs as well as the
other formats.
- Fix Lesstif issues of window placement and closing
Currently, when Lesstif is used, some of the default windows don't come
up in the correct positions and the Close buttons don't work properly.
Now uses OpenMotif. Done, for pave-3.0?
- Improved statistical capabilities
People often want to use simple statistics to assist with their data
analysis (e.g. mean, std. dev.). It would be good if PAVE could produce
these. In addition, people may want to slice the data in different ways to
obtain these statistics (e.g. mean for each time step or for each vertical
layer). There are also a number of commonly used statistics that deal with
comparing model data with observations. We may want to examine the "tables"
generated for OTAG for ideas for other typically used statistics.
- Properly visualize polar stereographic and other data
Only limited testing has been done using PAVE with the polar stereographic
map projection. More testing is required, and any bug fixes should be
implemented.
Done, for pave-3.0. Also added support
(as yet relatively un-tested) for Albers Equal Area and Transverse
Mercator projections.
- Add new types of plots and options for existing ones
Additional kinds of plots have been requested by users. Some examples
include: vertical cross section plots where the size of the layers is
proportional to actual layer size, vertical profiles, contour plots, flux
plots, observational data plots described above, PIG plots, nested grid plots,
adaptive grid plots, box & whisker plots. Also, making scatter plots internal
to PAVE would help reduce the problems associated with transferring data to
BLT and would provide additional flexibility.
- Improve use of map backgrounds
It would be useful to be able to plot multiple maps (e.g. state + counties,
or counties + roads) using different colors and thicknesses as needed.
Another possibility would be to allow backgrounds generated by packages such
as GISs to be displayed - these might show roads, water, cities, etc.
New maps addedfor pave-3.0.
- Increase configurability of plots
It would be useful in some cases to allow users to define the cutoffs
between the colors plotted (currently PAVE makes each bin of equal size from
the min to the max). Another alternative might include assigning bins according
to percentiles. Additional configuration options could be added to vector
plots wrt the density of the vectors plotted.
- Improve plot labeling & layout
The density and location of grid cell numbering on tile plots and in the
domain selector could be improved. Also, the legend could be outlined in black
to set off any white or light values and be positioned relative to the size of
the legend labels. The size of tile plots could be made user configurable,
and the aspect ratio preserved when the plot is manually resized. Data
plotted on time series plots could be labeled more clearly.
- Optimize PAVE performance and memory usage
PAVE performance, both CPU and memory related, can be improved in a number
of ways. For example, loops can be reordered to reduce paging to disk, memory
leaks can be plugged, and memory can be reused in some formulas.
Done, for pave-3.0.
- and many more...
Return To Table of Contents
Return To cjcoats.github.io
$Id: PaveManual.html 87 2018-03-14 13:34:05Z coats $
Send comments to
Carlie J. Coats, Jr.
carlie@jyarborough.com








